Kihara lab predicting assembly order of multimeric protein complexes
03-29-2018
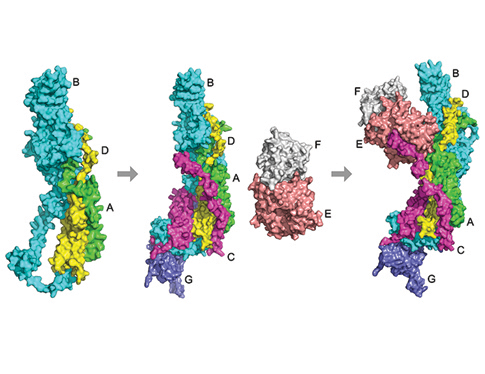
Protein-protein interactions, particularly those involving multiple proteins, are the cornerstone of numerous biological processes. Although an increasing number of multi-chain protein complex structures have been determined, fewer studies have been performed to determine the assembly order of complexes. Knowing the assembly order of a complex provides insights into the process of complex formation. Assembly order is practically useful for reconstructing and determining the structure of a subcomplex of a large protein complex. It also has important applications including designing artificial protein complexes and drugs that prevent the assembly of protein complexes.
Our lab has developed a computational method, Path-LZerD, which predicts the assembly order of a protein complex by simulating its assembly process. This is the first method of this kind. A strong advantage of Path-LZerD is that the assembly order can be predicted even when the overall complex structure is not known. Path-LZerD opens a new area of computational protein structure modeling and will be an indispensable approach for studying protein complexes.
This work was published recently on PLOS Computational Biology:
http://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.1005937
Modeling the assembly order of multimeric heteroprotein complexes. Lenna X. Peterson, Yoichiro Togawa, Juan Esquivel-Rodriguez, Genki Terashi, Charles Christoffer, Amitava Roy, Woong-Hee Shin, & Daisuke Kihara, PLoS Comput. Biol. (2018), 14:e1005937
Contact: Daisuke Kihara http://kiharalab.org