Gelvin lab identifies epigenetic process impacting Agrobacterium-mediated transformation of Arabidopsis
12-15-2016
Name of paper:
Iwakawa, H., Carter, B.C., Bishop, B.C., Zhu, Y., Ogas, J.,and Gelvin, S.B. 2017. Perturbation of H3K27me3-associated epigenetic processes increases Agrobacterium-mediated transformation. Mol. Plant-Microbe Interact. doi.org/10.1094/MPMI-12-16-0250-R. In press.
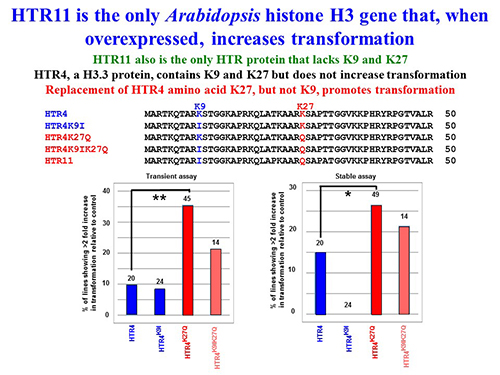
Figure legend:
Transient and stable transformation of Arabidopsis lines overexpressing HTR4 variants. (A) Top, amino acid sequences of histone H3-11, H3-4, and H3-4 variants. Only the N-terminal sequences of the proteins are shown; bottom, schematic maps of the constructs used for histone overexpression. 2x35S promoter, Cauliflower Mosaic Virus double 35S promoter; nos terminator, nopaline synthase terminator; (B) Transient transformation of T2 generation Arabidopsis transgenic lines overexpressing the indicated HTR4 or HTR4-variant cDNAs.