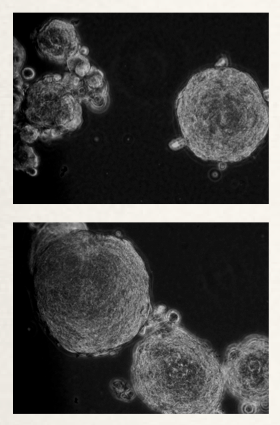
Figure 1: KRAS mutant, untransformed human bronchial epithelial cells growing in soft agar following transduction with a miRNA library. Ability to grow in soft agar suggests that the cells have undergone transformation. Clones are harvested and the miRNAs responsible for the transformed phenotype are identified. Follow-up cell-based and in vivo studies confirm the transformation potential of the identified miRNAs.
Project 3:
Identification of miRNAs that potentiate KRAS- and/or p53- driven lung tumorigenesis
This project will identify miRNAs that can potentiate tumors with activated KRAS and/or mutant p53 (Figure 1). Techniques to determine miRNA involvement in oncogenesis have historically been done through deep sequencing or microarrays of patient-derived tissue. These data sets, while valuable for correlative studies, do not supply direct evidence for individual miRNA involvement in promoting tumorigenesis. Without this specific data moving forward with miRNA-based therapeutics is done in a blind fashion. Therefore the significance of this project is to separate those miRNAs that are relevant to the tumorigenic process from those with secondary effects, specifically in the background of the KRAS oncogene and/or in combination with mutant p53. Filtering out the noncausal miRNAs will allow future therapeutic studies to focus on the cancer-promoting miRNAs that have clinical relevance. This knowledge will also point to miRNAs that should be further evaluated and targeted in the clinic.