Colicins. Colicins are plasmid-encoded protein antibiotics that target E. coli. Most Gram-negative bacteria produce such bacteriocins. Colicins exert their lethal effect on sensitive E. coli cells, which do not contain a protective immunity protein through: (i) intracellular nuclease action on DNA, rRNA, or tRNA, (ii) formation of depolarizing and de-energizing ion-conductive channels in the cytoplasmic membrane, or (iii) inhibition of cell wall murein synthesis. Studies on the mechanisms of action of nuclease colicins and pore-forming colicins have contributed to increased understanding of ribosome structure-function, protein-membrane interactions, and protein import.
Crystal structures of colicins; BtuB and OmpF Receptors. As for many toxins, such as diphtheria, colicins are multi-domain structures. Colicins contain three functional and structural domains, the N-terminal T- (translocation), central R- (receptor binding), and C-terminal C (cytotoxic) -domains (green, red, and blue, Fig. 1A). These domains function in translocation (T) of the colicin across the outer membrane, initial tight binding to a receptor (R) in the outer membrane, and in cytotoxicity (C) by the pore-forming or degradative enzymatic mechanisms discussed above. Structures are shown of four colicins [colicin B, E3, Ia, and N] (Figs. 1B-E). The tightly wound alanine-rich long coiled-coil (i. e., “ala-coil”) of the receptor (R) binding domain seen prominently in the structures of colicins E3 and Ia, 100 Å and 160 Å, respectively, is particularly striking. We have shown that the extended length of the R domain contributes to the structure of an outer membrane translocon by connecting through lateral diffusion in the outer membrane, the BtuB receptor and OmpF translocator proteins.
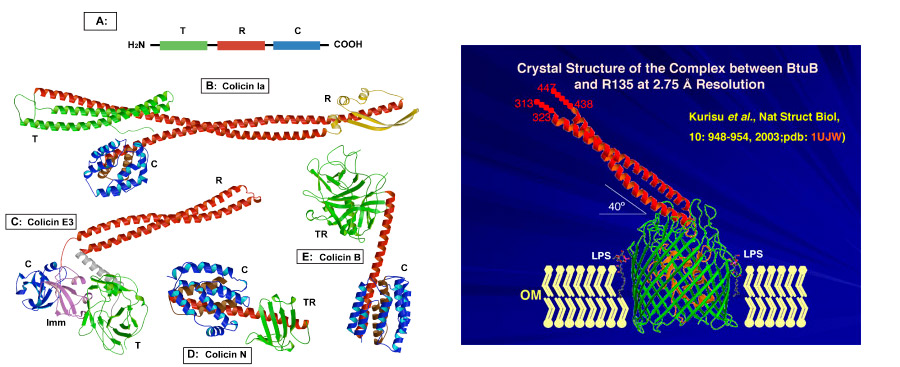
Fig. 1, left:. Colicin structures obtained elsewhere (A) Colicin domain structure; ribbon diagrams of crystal structures of colicins Ia,(B).E3, (C).N, (D), and B (E). (F , right ) Crystal structure (2.75 Å) of the complex between the vitamin BtuB receptor and the 135 residue receptor-binding domain (R135) of colicin E3 (Kurisu et al., 2003).
Structure-Function of OmpF. OmpF is one of three trimeric outer membrane porins, including OmpC and PhoE, whose principle physiological function is to allow passage through the OM of small, hydrophilic nutrient molecules. OmpF was the first integral membrane protein from which crystals were obtained that diffracted to a meaningful resolution. The first crystal structure of OmpF diffracted to 2.4 Å, and the highest resolution that had previously been attained before the present studies was 2.2 Å.
Colicin import; the Nuclease Colicins. The problem of colicin translocation across the E. coli double membrane provides a system for studies of import mechanisms that has the advantage of an easily identifiable marker for completion of import - death of the target cell.
The outer membrane translocon. In contrast to the studies carried out thus far on organelle import, it has been possible to visualize segments of the imported colicin protein bound to both the colicin receptor, BtuB (Fig. 1F) and inserted into OmpF, which was proposed to be the translocator. Following the initial tight binding of the colicin R-domain to the BtuB receptor (Fig. 1F), the long coiled-coil R domain can to “fish for” and recruit the OmpF or OmpC, the T-domain inserts into and is anchored by OmpF, and the C-domain of the colicin can insert into and be transferred through another pore of the same OmpF molecule. A hypothetical model for the OM translocon (Fig. 2) followed from the original BtuB-R135 structure. At the time of the initial formulation of this model, there were no biochemical or biophysical data to support the involvement of OmpF.
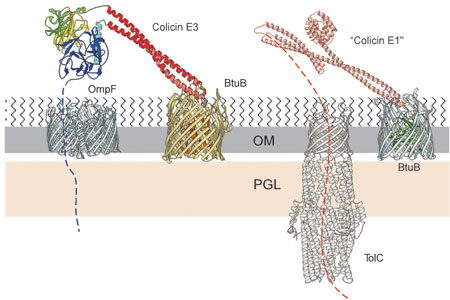 Fig. 2. Model for a two component translocon used involving the outer membrane receptor, BtuB translocator OmpF, and receptor-translocator, TolC, used for import of colicins E3 and E1 through the outer membrane. The colicin bound to BtuB shows the positions of the colicin R135 receptor-binding (red), translocation (blue), catalytic (magenta), and immunity (yellow) domains based on, and extrapolated from, the structure of the R135-BtuB complex [Fig. 1F], and a neighboring OmpF trimer. Orientation of T, C, and Imm domains is based upon the reference R domain and alignment of its BtuB receptor in the membrane. The threading of the colicin N-terminal T domain through a pore of the OmpF was hypothetical at that time, as were subsequent interactions with the TolB and Pal components bound, respectively, in the peptidoglycan (PG) layer and outer membrane.
Fig. 2. Model for a two component translocon used involving the outer membrane receptor, BtuB translocator OmpF, and receptor-translocator, TolC, used for import of colicins E3 and E1 through the outer membrane. The colicin bound to BtuB shows the positions of the colicin R135 receptor-binding (red), translocation (blue), catalytic (magenta), and immunity (yellow) domains based on, and extrapolated from, the structure of the R135-BtuB complex [Fig. 1F], and a neighboring OmpF trimer. Orientation of T, C, and Imm domains is based upon the reference R domain and alignment of its BtuB receptor in the membrane. The threading of the colicin N-terminal T domain through a pore of the OmpF was hypothetical at that time, as were subsequent interactions with the TolB and Pal components bound, respectively, in the peptidoglycan (PG) layer and outer membrane.
Import of the pore-forming colicin E1. From the requirement of the TolC outer membrane protein along with BtuB for cytotoxicity of colicin E1, a similar model for import of colicin E1 was proposed in which TolC, which spans the E. coli cell envelope, from the inner membrane across the periplasmic space through the outer membrane, serves as the translocator for colicin E1. Its outer membrane translocon is proposed to consist of BtuB/colicin E1/TolC or TolC alone (Fig.2).
The colicin E1 channel domain; channel formation. The soluble active (channel-forming) 20 kDa C-terminal domain of colicins E1 (Elkins et al, 1994; pdb, 2I88) and A, as well as the C-terminal domain of colicin Ia, has a characteristic 10-helix structure, which is similar to the structure of the T-domain of diphtheria toxin and the apoptotic protein, BclXL. The single channel conductance of the colicin channel (~ 106 ions/sec in 0.1 M ionic strength) is sufficient to depolarize the E. coli cell. The requirements for channel formation include a pronounced α-helical content of the channel domain bound to the surface of the membrane, mobility of the two dimensional helical array on the membrane surface, and the presence of anionic lipid in the membrane, so that channel formation in synthetic lipid membranes has a pronounced dependence on the anionic lipid content and surface potential, ?o, of the target membranes (Fig. 3) .
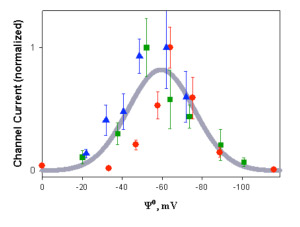
Fig. 3. Dependence of channel activity of colicin E1 channel domain on membrane surface potential. Macroscopic channel conductance measured at 25 mol % (red symbols), 30 % (green), and 70 % (blue) at 0.1 M, 0.3 M, and 1.0 M ionic strength, respectively (24).
Import of Nuclease Colicins; Structures. The steps 1-6 (Fig. 4) that can be defined in import of the nuclease colicins are: [1] import is initiated by the binding of colicin through its R-domain to the BtuB receptor. The existence of this tightly bound complex is documented by the crystal structures shown in Figs. 1F and 6A, B. The sequence of the subsequent minimum set of necessary events 2-6 in the import process is not known: [2, 3] binding of 83 residue disordered T-domain (for colicin E3) to OmpF and subsequent insertion of N-terminal domain containing the TolB interactive ‘box’ to allow interaction with TolB; [2] unwinding of R-domain coupled to unfolding of N-terminal T- and C-domains and [4] release of tightly bound immunity protein; [6] proteolytic cleavage of the ~ 96 residue C-domain to liberate it for passage through the OmpF pore (Fig. 5).
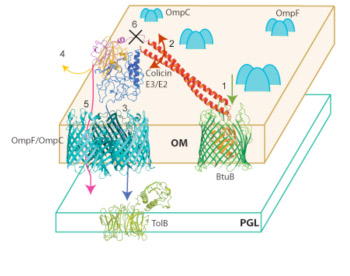
Fig. 4 Model for import of colicin E3 through the outer membrane utilizing the BtuB-OmpF translocon; OmpF is abundant in the OM, ~ 105 copies/cell. The alternative possibility of using OmpC (109) is indicated. Defined events in import are [1] binding of colicin to BtuB receptor; [2, 3] binding/insertion of T-domain to/into OmpF; [4] release of Imm protein and separation/unfolding of T- and C-domains; [5, 6] proteolytic cleavage between R and C domains to allow release of unfolded C-domain and [5, 6]insertion through OmpF pore into the periplasmic space.
The model shown in Fig. 4 was inferred from the following structure-function studies of the BtuB-OmpF colicin import system.
Step 1. Binding to BtuB; crystal structures of BtuB/E3-R135 and BtuB/E2-R135. Insight into the nature of the requirement resulted from the 2.75 Å crystal structure (Fig. 1F) of the vitamin B12 (BtuB)/E colicin receptor with the 135 residue coiled-coil receptor binding domain of colicin E3. Although bound predominantly to the extracellular loops of the BtuB receptor, the R135 is tightly (Kd <= 10-10 M) bound. It does not penetrate deeply into the receptor, making contact only with the outer surface of the core “plug” domain (green sub-structure shown in Fig. 1F), and does not displace the plug or change its structure. These structure data implied that BtuB is used only for binding and localization of the colicin on the outer membrane surface, and that the colicin must use a second OM receptor-translocator for translocation.
The structure of R135/BtuB complex, including the details of the R135 –BtuB interaction and the oblique orientation of R135, has now also been shown (3.5 Å resolution) for the DNAase colicin E2 (Fig. 5A), showing that it is unlikely that the oblique nature of the extended receptor-bound colicin R-domain, which would facilitate interaction interaction with OmpF (Fig. 4) could be a consequence of crystal contacts. Furthermore, although the colicin R-domain is involved in crystal contacts, all of the crystal contacts of the E3 R-domain with BtuB in the two structures are with the flexible loops 5-6 and 7-8 of BtuB. These loops are highly flexible as implied by extracellular loop 5-6 being disordered in both the in surfo and in meso BtuB structures and loop 7-8 disordered in the in surfo structure.
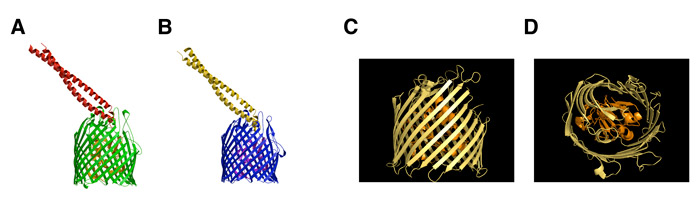
Fig. 5A, B. Ribbon diagrams of the complex of the 135 residue receptor binding domain of colicins E2 (left ) and E3 (right), the latter structure described in (1). (C) Ribbon diagram of the 1.95 Å structure of BtuB obtained in collaboration with V. Cherezov and M. Caffrey.
Further structure studies on BtuB; crystallization in meso (lipid cubic phase) of BtuB resulting in a 1.95 Å resolution structure. Using short chain monoacylglycerols as bilayer hosts in a cubic phase for growing diffraction-quality crystals of membrane proteins, in collaboration with V. Cherzov and M. Caffrey, a higher resolution structure (1.95 Å) of BtuB was determined (Fig. 5C) than had been obtained in surfo. BtuB is the first integral membrane protein, which is not rhodopsin-like, to be analyzed by crystallization in the lipid cubic phase (in meso) and to provide better diffraction than was obtained previously from crystals made in detergent (in surfo). The second such ‘different’ protein crystallized in meso was the ?-adrenergic receptor. An interesting finding that is fundamental to the structural biology of membrane proteins is that there are significant differences in the structure of BtuB determined in meso relative to that determined from crystals formed in detergent (in surfo): (i) the conformation of residues 6-7 in the TonB box, considered critical to signal transduction, is completely different. (ii) The five N-terminal residues of the TonB box are more ordered; (iii) residues 57-62 in the hatch domain are more disordered; (iv) extracellular loops 9-10, 13-14, 15-16, 19-20 have a different position, as do residues 86-96 in the hatch domain.
Step 2 : separation, unfolding, of the C- and T- domains. During the binding and insertion interactions with the outer membrane proteins, the C- and T- domains must be separated, unfolded, and released. The only known source for free energy to accomplish this separation and unfolding would be the energy of binding to BtuB and any other OMP component of the translocon. Colicin E3 could be labeled with a fluorescence energy donor and acceptor, a “FRET pair”. Separation of the T- and C-domains was assayed in vitro through the decrease in FRET efficency upon binding to purified BtuB or OmpF, which implies an increase in the distance of separation of T- and C-domains.
Step 3 (Fig. 5): Binding of colicin E3 to OmpF assayed through occlusion of OmpF ion channels. It is not possible to measure the relatively weak binding of colicin E3 to OmpF by microbiological spot tests, as can be done to characterize the tight binding of the colicin to BtuB. Alternatively, qualitative aspects of colicin - OmpF binding can be measured by utilizing the ability of OmpF to form highly conductive ion channels. The OmpF single channel conductance is 1 nano-Siemen in 1M ionic strength. Colicin E3 can occlude OmpF channels when it is added to the trans (Fig. 7B), but not cis (Fig. 7A), side relative to the side of OmpF addition to the planar bilayer. For intact OmpF, the additional requirement for occlusion is a membrane potential that is trans-negative relative to the side of addition of the colicin. These condition are summarized in the histogram of Fig. 7E, where it is seen that the channels are completely occluded when E3 is added trans and the potential is -50 mV cis. Once in the bilayer the occlusion by the colicin is irreversible as it cannot be reversed by perfusion (Fig. 6C).
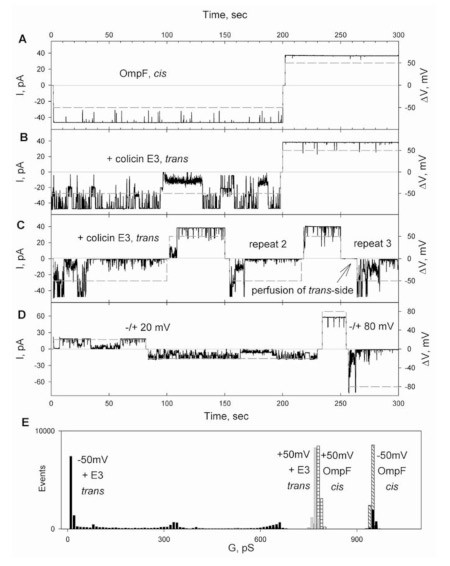 Fig. 6. Colicin occlusion of OmpF channels in planar bilayers. OmpF, 10 pg/ml, added from cis compartment. (A) Trans-membrane current (solid) through one OmpF trimer; (B) Colicin E3, 50 ng/ml, added to trans-side; 500 ng/ml; (D) Effect on occlusion and opening of channels on size of TM potential; (E) Histogram of OmpF channel conductance, dependence on polarity of TM potential in the absence and presence of trans-side colicin.
Fig. 6. Colicin occlusion of OmpF channels in planar bilayers. OmpF, 10 pg/ml, added from cis compartment. (A) Trans-membrane current (solid) through one OmpF trimer; (B) Colicin E3, 50 ng/ml, added to trans-side; 500 ng/ml; (D) Effect on occlusion and opening of channels on size of TM potential; (E) Histogram of OmpF channel conductance, dependence on polarity of TM potential in the absence and presence of trans-side colicin.
Occlusion by the unfolded T-domain. The 83 residue N-terminal segment of colicin E3 was shown in the original crystal structure to be unfolded. It is particularly potent in occluding the OmpF channels as its occlusion does not require a trans-membrane potential.
Specificity of occlusion. Colicin E1, which is known not to interact with OmpF, but with TolC along with BtuB, occludes channels formed by TolC, but not OmpF.
Steps 4, 5 Release of immunity protein; Requirement of unfolded C-domain for occlusion of OmpF channels. The 96 residue endoribonucleolytic C-domain isolated in its folded form while bound to immunity, does not occlude OmpF channels. Using a novel procedure for removal of immunity protein, high affinity anion exchange, the immunity protein was released and, concomitantly, the C-domain assumed an unfolded state as determined by its far-UV circular dichroism spectrum. The Imm-free unfolded cytotoxic C-domain was active in occlusion of the 80 picoSiemen OmpF channels.
Occlusion of OmpF by unfolded C-domain. Ultimately, it is of course the C-terminal active domain (the 96 residue colicin fragment, ‘C96’; blue domain in Fig. 1C) that must be inserted through the outer membrane in order to exert its degradative or dissipative influence in the cytoplasm or inner membrane. It was found that C-terminal domain (C96) that was folded in the presence of the tightly bound immunity protein (Kd ? 10-13 M between colicin E3 and Imm, and 10-11 M between C-domain and Imm) did not occlude OmpF. A detailed structure of the colicin E3-Imm structure, shown in a ribbon diagram magnified from the version presented in Fig. 1C is shown (Fig. 7).
The relatively simple procedure developed for removal of Imm from the colicin or C96 without denaturation by passage of purified His-tagged C96 through a Ni2+ column or of colicin E3 though a high affinity anion exchange column (HiTrap Q, Amersham). Apparently, the previous inability to remove Imm without denaturation of the colicin was partly a consequence of the prior use of more weakly binding anion exchange columns.
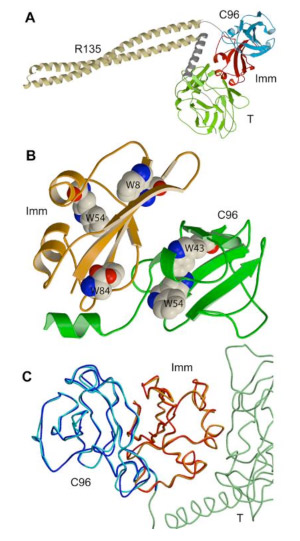 Fig. 7 (left). Ribbon diagram of crystal Structure of colicin E3 with its immunity protein. (A) Magnified version of Fig. 1C; (B) Position of Trp side chains that are useful as fluorescence probes and for FRET analysis; (C) Superposition of backbones of C96 and Imm.
Fig. 7 (left). Ribbon diagram of crystal Structure of colicin E3 with its immunity protein. (A) Magnified version of Fig. 1C; (B) Position of Trp side chains that are useful as fluorescence probes and for FRET analysis; (C) Superposition of backbones of C96 and Imm.
Two structures of the OmpF porin: (I) High (1.6 Å) resolution structure of the OmpF porin. While screening crystallization conditions in order to obtain a complex of OmpF and T83, a 1.60 Å data set was obtained of the E. coli OmpF porin. The best data set presently in the pdb for the E. coli OmpF porin, which was first crystallized in 1980, has a resolution of 2.2 Å (pdb: 1HXX). The 1.6 Å OmpF structure (pdb: 2ZFG, pre-publication) was obtained by co-crystallization of OmpF in the presence of a high concentration (1 M) of MgCl2 and the glycine-rich 83 residue N-terminal domain of colicin E3 that is disordered in the crystal structure of the intact colicin E3. The arrangement of the backbone and side chains of OmpF determined in the present study is not changed significantly relative to the 2.2 Å structure (Fig. 2E) previously determined. The rmsd between the present 1.6 Å and the 2.2 Å structure is 0.26 Å.
The Fo–Fc difference map in the 1.6 Å OmpF structure shows the presence of additional electron density of a (H2O)6 - coordinated Mg2+ ion (orange) near Asp113, Leu115, and Glu117 in the L3 loop of the selectivity filter (Fig. 8A) and the positively charged array Arg42, Arg82, Arg132, Lys16, Lys80, and Glu62 on the opposite side of the limiting filter aperture. The cross-section of the limiting aperture of the selectivity filter is: 7.8 Å (Arg82NH - Asp113OD2) – 8.8 Å (Lys16NZ - Leu115O) x 14.4 Å (Tyr310OH - Tyr106OH, or Tyr310OH - Tyr102OH). The solvent accessible cross-sectional area subtended by these distances across the limiting channel aperture is elliptically shaped, with short and long axes of approximately 7.8 Å x 13 Å, slightly larger than the 7 x 11 Å cross-section derived from the original OmpF structure that is cited in much of the OmpF literature. The hydrogen-bonding pattern of the (H2O)6 - Mg2+ to Asp113OD1 and to the backbone carbonyls of Leu115 and Glu117 is shown (orange, Fig. 8B). The decrease in cross-sectional area of the restriction zone caused by the presence of the bound hexa-Mg2+ ion resulted in an approximately proportional decrease in the conductance of OmpF as a function of ionic strength in the presence of a low concentration (33 mM) of MgCl2 (Fig. 8C).
Steps 3 and 5 ). Demonstration of a colicin peptide inserted into OmpF. (II) Structure of the OmpF porin containing a glycine-rich peptide segment of the N-terminal T83 of colicin E3. It has been possible to obtain crystals of a complex of OmpF and T83 using a strategy opposite to that employed to obtain the OmpF crystals that diffracted to 1.6 Å. This strategy is to form the complex and carry out the crystallization under conditions of very low ionic strength. The co-complex can be isolated by size-exclusion chromatography with a stoichiometry of approximately 1 peptide: OmpF. Crystals of the putative complex diffracted to 3.0 Å. A ribbon diagram of the complex shows a difference (Fo- Fc) electron density (blue) that defines a peptide (Fig. 9) that extends through the selectivity zone defined by loop 3. The end of loop 3, also seen in Fig. 8A, contains a short helical sequence, shown in red in Fig. 16. The peptide is rich in glycine. T83 contains 34 Gly in its 83 residues.
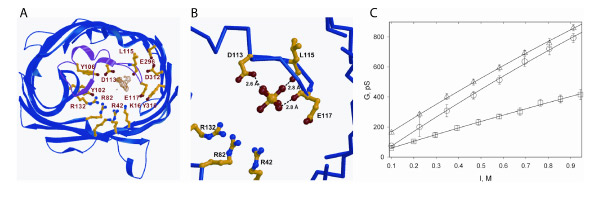
Fig. 8 (A) Fo–Fc difference map in the 1.6 Å OmpF structure showing the electron density of a hexa-H2O coordinated Mg2+ atom (orange) near Asp113, Leu115, and Glu117 in the L3 loop of the selectivity filter. Also shown are Arg42, Arg82, Arg132, Lys16, Lys80, and Glu62 on the opposite side of the limiting aperture. (B) H-bonding of (H2O)6 - Mg2+ to Asp113OD1 and the backbone carbonyls of Leu115 and Glu117. (C) Single channel conductance of OmpF in planar bilayer membranes as a function of ionic strength (I): NaCl (triangles), NaCl in the presence of 33 mM MgCl2 (circles); MgCl2 (squares).
Two structures of the OmpF porin: (I) High (1.6 Å) resolution structure of the OmpF porin. While screening crystallization conditions in order to obtain a complex of OmpF and T83, a 1.60 Å data set was obtained of the E. coli OmpF porin. The best data set presently in the pdb for the E. coli OmpF porin, first crystallized in 1980, has a resolution of 2.2 Å (pdb: 1HXX). The 1.6 Å OmpF structure (pdb: 2ZFG, pre-publication) was obtained by co-crystallization of OmpF in the presence of a high concentration (1 M) of MgCl2 and the glycine-rich 83 residue N-terminal domain of colicin E3 that is disordered in the crystal structure of the intact colicin E3 (40). The arrangement of the backbone and side chains of OmpF determined in the present study is not changed significantly relative to the 2.2 Å structure (Fig. 2E) previously determined. The rmsd between the present 1.6 Å and the 2.2 Å structure is 0.26 Å. The Fo–Fc difference map in the 1.6 Å OmpF structure shows the presence of additional electron density of a (H2O)6 - coordinated Mg2+ ion (orange) near Asp113, Leu115, and Glu117 in the L3 loop of the selectivity filter (Fig. 8A) and the positively charged array Arg42, Arg82, Arg132, Lys16, Lys80, and Glu62 on the opposite side of the limiting filter aperture. The cross-section of the limiting aperture of the selectivity filter is: 7.8 Å (Arg82NH - Asp113OD2) – 8.8 Å (Lys16NZ - Leu115O) x 14.4 Å (Tyr310OH - Tyr106OH, or Tyr310OH - Tyr102OH). The solvent accessible cross-sectional area subtended by these distances across the limiting channel aperture is elliptically shaped, with short and long axes of approximately 7.8 Å x 13 Å, slightly larger than the 7 x 11 Å cross-section derived from the original OmpF structure (51) that is cited in much of the OmpF literature (50). The hydrogen-bonding pattern of the (H2O)6 - Mg2+ to Asp113OD1 and to the backbone carbonyls of Leu115 and Glu117 is shown (orange, Fig. 8B). The decrease in cross-sectional area of the restriction zone caused by the presence of the bound hexa-Mg2+ ion resulted in an approximately proportional decrease in the conductance of OmpF as a function of ionic strength in the presence of a low concentration (33 mM) of MgCl2 (Fig. 8C).
The high concentration of MgCl2 and the presence of the T83 N-terminal fragment of colicin E3 were special conditions for crystallization. It had been believed that high ionic strength facilitated insertion of the T83. This assumption turned out to be erroneous and T83 was not seen in the structure, although a unique Mg2+ ion was present (Fig. 8B). The value of the higher resolution OmpF structure derives from the extensive use of OmpF as a model for theoretical and molecular dynamics studies of the forces that guide ion flow in trans-membrane channels (54, 126-130). Ordered waters in the channel affect the magnitude and distribution of the electrostatic forces within the channel.
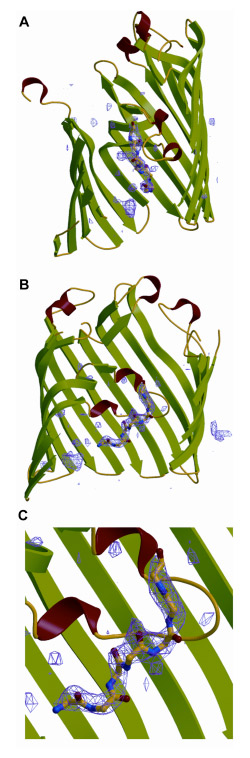
Fig. 9. (A, B) Panels differing by a 90° rotation around the pore axis that describe the F0 minus FC map of OmpF and additional electron density attributed to a terminal region of the T83 peptide. The additional density consists of at least seven residues and extends over a distance of at least 18 α. Top, extracellular, and bottom, periplasmic side of OmpF. (C) Region of interaction of T83 peptide and loop 3 of OmpF shown in enlarged format.
Summary of crystal structures obtained in these studies. The crystal structures from structure studies obtained in studies on the problems of colicin-membrane interactions and cellular colicin import, which have been deposited in the protein data bank, are: (i) the 190 residue C-terminal domain of colicin E1 (pdb: 2I88); in collaboration with C. V. Stauffacher; (ii) complex of colicin E3 receptor binding domain and BtuB (pdb:1UJW); (iii) BtuB in meso (2GUF); with V. Cherezov and M. Caffrey. (iv) complex of colicin E2 receptor binding domain and BtuB (pdb: 2YSU); (v) OmpF,1.6 Å resolution (pdb, 2ZFG; pre-publication) (vi) OmpF with inserted T83 fragment of colicin E3, 3.0 Å (rdb, 2ZLD, pre-publication).
Studies on α-Synuclein. Experimental approaches used to define channel properties of colicin E1, a combined spectroscopic and electrophysiological approach were applied to ?-synuclein, a 14.5 kDa protein associated with presynaptic nerve terminals and neuronal vesicles, which is implicated in the etiology of Parkinson’s disease. Intrinsically disordered in aqueous environment, monomeric aS develops a highly helical conformation upon binding to membranes containing (i) anionic lipids; (ii) PE lipid with a tendency to support negative curvature in membranes and thereby non-bilayer structures, and (iii) membranes vesicles of small diameter. The consensus in the literature was that the membrane-permeabilizing acticity of α occurred through oligomeric ‘protofibrillar’ forms that have a predominantly α-strand secondary structure.
In our studies carried out with C. Rochaix, monomeric wild type α and two mutants associated with familial PD, E46K and A53T, formed ion channels with well defined conductance states in membranes containing 25-50% anionic lipid and 50% phosphatidyl-ethanolamine (PE) in the presence of a trans-negative potential. Another familial mutant, A30P, known to have a lower membrane affinity, did not form ion channels. Ca2+ prevented channel formation when added to membranes before α, and decreased channel conductance when added to preformed channels. In contrast to the monomer, membrane permeabilization by oligomeric α was not characterized by formation of discrete channels, or a requirement for PE lipid or a membrane potential. In contrast, monomeric wild type α and two mutants associated with familial PD, E46K and A53T, formed ion channels with well defined conductance states in membranes containing 25-50% anionic lipid and 50% PE in the presence of a trans-negative potential. Another familial mutant, A30P, known to have a lower membrane affinity, did not form ion channels. Ca2+ prevented channel formation when added to membranes before α, and decreased channel conductance when added to preformed channels.
Channel activity, α-helical content and thermal stability of membrane-bound α determined by far-UV CD, and lateral mobility of α bound to planar membranes measured by fluorescence correlation spectroscopy, were correlated. It was inferred that discrete ion channels with well defined conductance states were formed in the presence of a membrane potential by several molecules of α-helical monomeric α. Such channels may have a role in the normal function and/or physiology of the protein. One possible function would be as a channel or transporter, of dopamine, a biogenic amine neurotransmitter.
 © William Cramer Lab
© William Cramer Lab