Research
Video Overview, Superheroes of Science:
Discovering proteins and searching for a treatment to Parkinson’s
https://www.youtube.com/watch?v=pNMeXiJY-VI
What are Fic (Filamentation induced by cAMP) proteins?
Fic proteins derive their name from a cell division (filamentation) phenotype attributed to mutations in the fic gene of E. coli in response to cAMP.
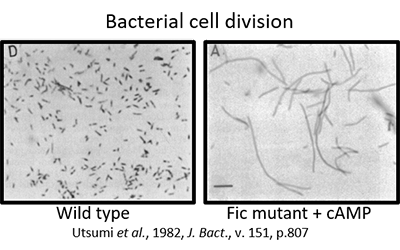
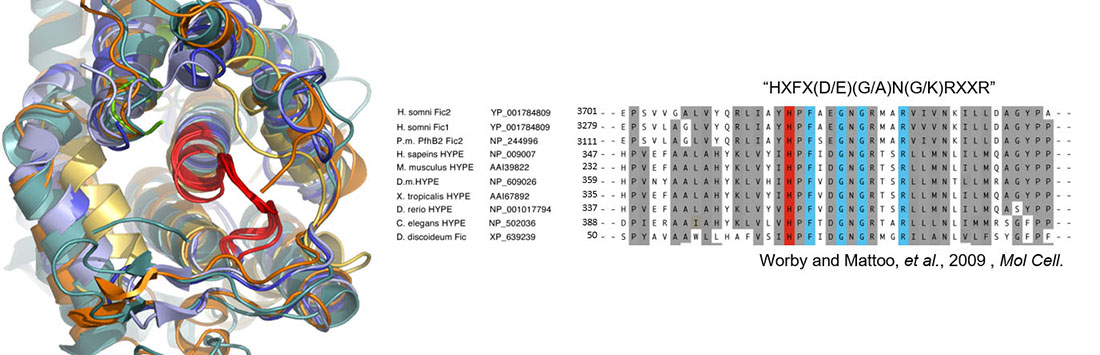
Fic proteins are defined by a conserved HxFx(D/E)(G/A)N(G/K)RxxR sequence within an alpha-helical protein fold. The invariant Histidine is required for enzymatic activity.
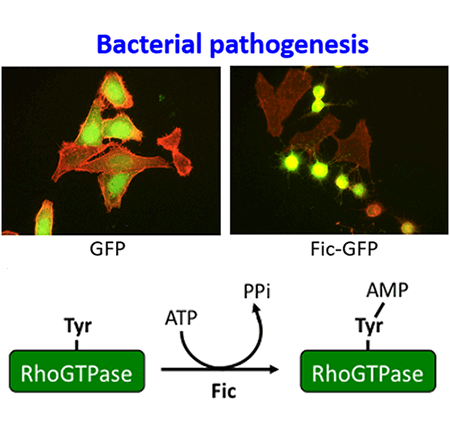
What do Fic proteins do?
We showed that pathogenic bacteria could secrete Fic proteins to AMPylate and inactivate mammalian Rho GTPases, thereby inducing cytotoxicity. This is a mechanism by which bacteria evade the host’s immune response.
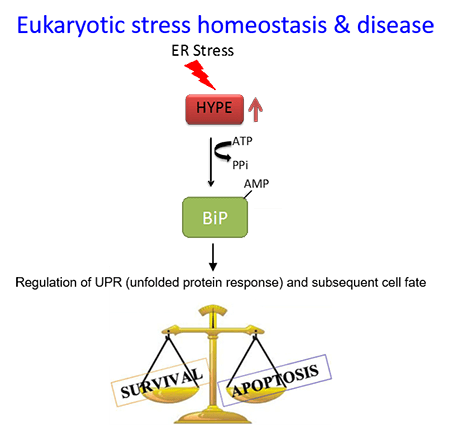
We also showed that Fic-mediated adenylylation is conserved in humans. Specifically, we discovered that the human Fic protein, HYPE, AMPylates the Hsp70 chaperone, BiP, in the lumen of the endoplasmic reticulum (ER). In so doing, HYPE alters how eukaryotic cells respond to ER stress via a pathway called the unfolded protein response (UPR). Thus, HYPE was established as a novel regulator of ER homeostasis and an important determinant of cell fate.
What does our lab do?
The Mattoo Lab takes a multidisciplinary approach to studying the functional repertoire of Fic proteins in prokaryotic and eukaryotic signal transduction. We follow interesting and evolutionarily distinct Fic proteins (eg. HYPE, IbpA, PfhB2, EcFic2, BbFic, Doc, etc.), assess their enzymatic activity, and identify their targets, with a larger goal of deciphering their cellular functions. Importantly, such an approach has allowed us to open new avenues of scientific research.

