Flaviviruses: Dengue virus, Yellow fever virus, West Nile virus, Zika virus
Replication and assembly
Replication of flaviviruses takes place on induced vesicles derived from the ER and requires the viral nonstructural (NS) proteins. We are investigating the functions of the nonstructural proteins that are involved in replication such as NS1, NS3 and NS4B. NS1 is a lipid-binding protein that is required for replication but the mechanism remains unclear. The structure of NS1 was recently determined and has guided a mutation-based analysis of NS1 function. NS3 is an enzymatic protein that acts as a protease and helicase. NS4B is a membrane protein that anchors the replication complex to the ER membrane. We are also interested in how replication is coupled with assembly and are using electron microscopy to determine the sequence of the events that drive virion budding, fission and trafficking.
Lipidomics, proteomics and metabolomics
We are investigating the interaction between cellular lipid metabolism and flavivirus infection. Sphingolipids are key regulators of cellular processes such as apoptosis, proliferation, cell migration and autophagy. They are also the major component in lipid rafts, which are important for viral entry. We are using inhibitors and siRNA to knock-down different lipid biosynthesis pathways and determine the effect on virus infection.
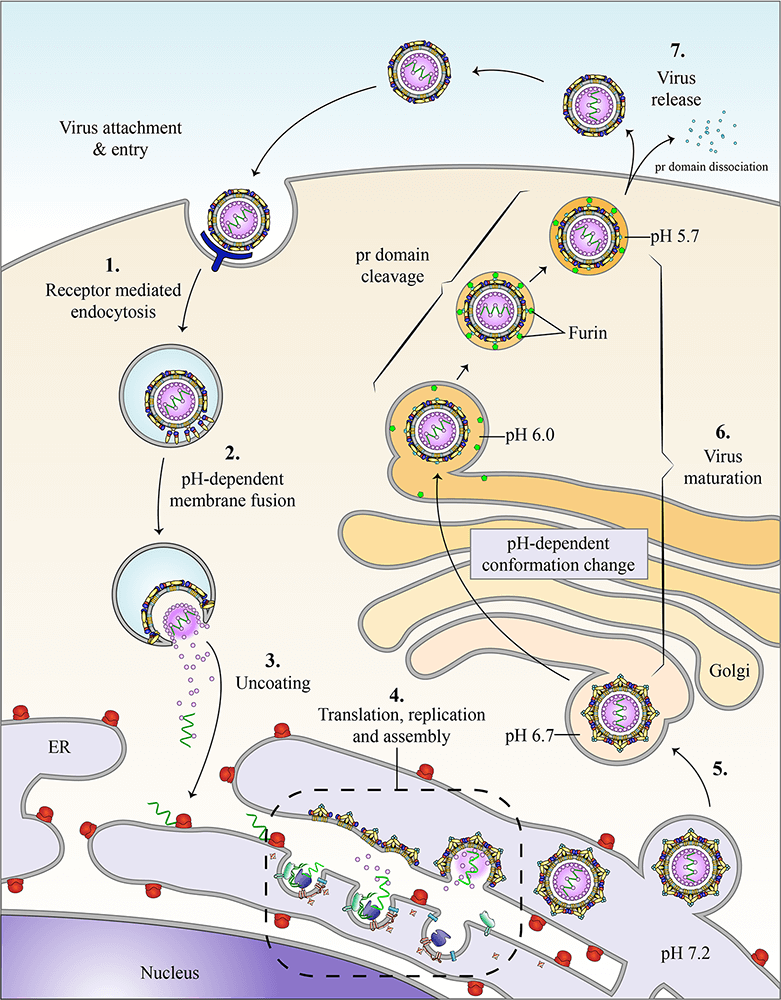
Viral maturation
Dengue virus is released from the host in a continuum of states, from fully immature virus to fully mature virus. We are using a combination of approaches to study the dynamics of the viral glycoproteins. We have proposed that the prM content (immaturity level) influences pathogenesis in humans, and we are interested in how virus maturity affects antibody neutralization, the immune response, and vaccine efficiency.
Alphaviruses: Sindbis virus, Chikungunya virus
Viral entry
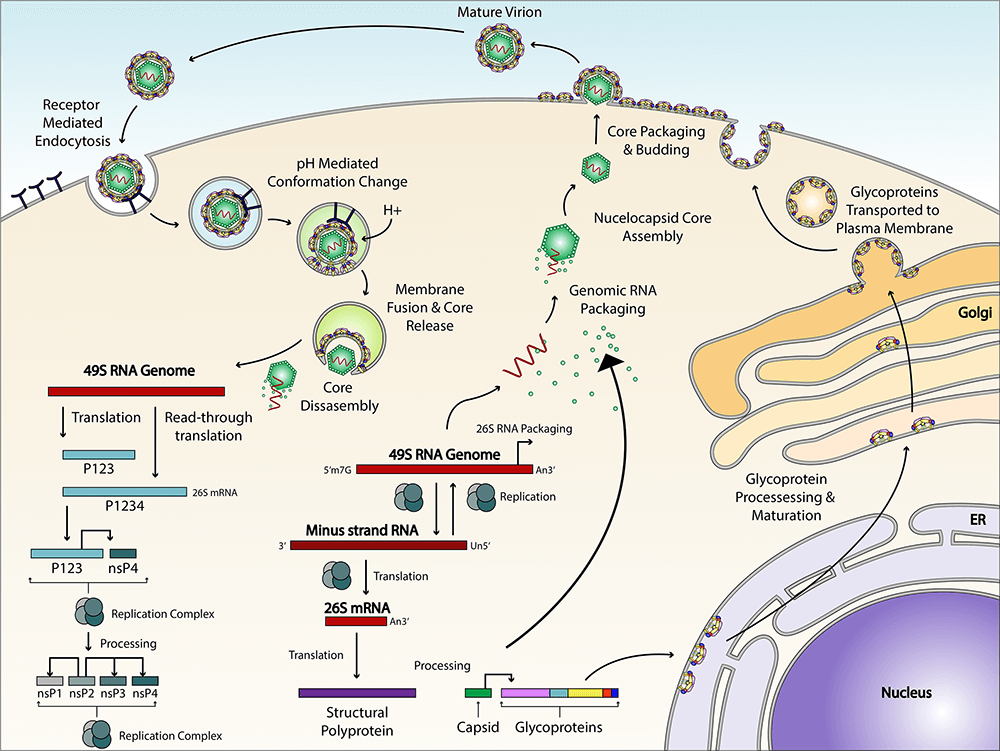
Alphaviruses enter the cell through receptor-mediated endocytosis. The low pH in the endosome triggers viral fusion and nucleocapsid is released into the cytoplasm. Before viral replication can begin, the nucleocapsid has to disassemble and release its RNA. It is believed that ribosomes facilitate this disassembly by interacting with capsid protein. We are using an in vitro assembly system to create nucleocapsid cores that can be tracked in live cells. We have previously made infectious virus-like particles by microinjecting nucleocapsid into cells expressing the viral glycoproteins. Using this system, we are interrogating the pathway for core disassembly and delivery of genomic RNA to the site of translation.
Replication
In alphavirus infected cells, the nonstructural proteins (nsPs) nsP1 to nsP4, along with host factors, form the cellular membrane-bound replication complex (RC) that performs the replication of the viral genome and transcription of the RNA. During virus budding the nucleocapsid cores that are assembled in the cytoplasm interact with the envelope glycoproteins at the plasma membrane to form virions. We use live imaging, biochemical, and structural tools to study the formation and assembly of viral RCs and particle budding in mosquito and mammalian cells.
Assembly and exit
RNA synthesis and capsid assembly are believed to be two separate and compartmentalized processes in alphaviruses and yet somehow viral RNA is delivered to a capsid molecule to initiate assembly. We are interested in protein and RNA compartmentalization and also how newly assembled nucleocapsid cores and other viral proteins use cytopathic vacuoles to traffic to the plasma membrane. We are using live cell imaging, super-resolution microscopy and electron tomography to visualize the sites of assembly and the viral transport system.