David Sanders Lab
Peer Review
One day in the late 1980s, I walked into the biochemistry laboratory at the University of California, Berkeley, where I was a PhD student, and declared that since Time magazine had a “man of the year” (it was only renamed “person of the year” later), Science magazine should have a “molecule of the year”. It happened that I was well positioned to make the suggestion: my supervisor, Daniel E. Koshland Jr, was the editor of Science.
The response to my proposal was the expected ridicule from my lab colleagues. Nevertheless, some time after I left the lab, Dan called me and said: “Guess what is going to be in the end-of-the-year issue of Science?” I guessed that it was something related to the lab’s research. “No,” he replied gleefully: “Molecule of the year!”
Far too much emphasis is placed on who is proposing to do the research and the institutions with which they are associated, rather than on the actual science. Therefore, I recommend that review be conducted in two stages. Reviewers should initially receive only descriptions of the proposed research, written in the third person, with no preliminary results section or indication of the authors’ identities or affiliations. This would require that the proposal be evaluated and scored solely on the detail of its merits.
The second step would see the reviewers provided with the usual biosketches, preliminary results, facility statements and so on, to help them evaluate the capacity of the researchers to conduct the proposed research. There can be no denial that the investigator’s track record is a relevant factor regarding a project’s potential for success. It is, however, inequitable if their identity or affiliation is the dominant factor in assessment.
The purpose of this two-part review is to reduce the biases, unconscious or conscious, that currently affect the evaluation of proposals.
Accountability could be further improved by attributing each review to its author. Some might object that confidentiality allows reviewers to be more honest, fearing retaliation less. In fact, confidentiality allows reviewers more scope to favour friends, retaliate against foes and exploit their privileged access to the information in the proposal to advance their own research programmes. The National Institutes of Health reports having detected examples of these forms of misconduct.
Peer review is a powerful method for evaluating research funding proposals. However, improving the fairness and accountability of peer review is a necessity if the modern scientific enterprise is to achieve the more equitable and wider distribution of resources necessary to fund innovative exploration.
Excerpted from a recent publication https://www.timeshighereducation.com/opinion/peer-review-should-be-two-stage-science-first-process
Scientific Publication Ethics
In academia, people are judged primarily on the basis of their output, which is most frequently in written form. It is, therefore, imperative that credit for the generation of that product be apportioned appropriately.
The US Federal Research Misconduct Policy defines plagiarism as “the appropriation of another’s ideas, processes, results, or words without giving appropriate credit”. So it is a violation to copy or to use a slightly modified version of another’s words without attribution – where attribution means not only citing the source of the text but also explicitly (by means of quotation marks or offsetting) indicating exactly which words are being reproduced. After all, academics could produce a much greater volume of writing if they didn’t have to actually craft their own text.
A more serious kind of academic plagiarism is the abuse of peer review. Reviewers are provided with privileged information on the condition that they will not share it with others and will not use it to advance their own scholarly activities. But readers of published articles often recognise duplicated texts, or may have strong reason to suspect that results described have been reproduced or materially influenced by a manuscript of their own that the author was asked to review.
Such behaviour is unfair and promotes the wrong concept of the route to academic success. Yet while students are held to account for plagiarism, academics rarely are. One major reason is that journal editors, the first line of defence in the battle against plagiarism, are highly reluctant to penalise plagiarism.
As for self-plagiarism, journals explicitly state that authors must not recycle their own writing – and certainly not without proper citation. It is not fair to those who follow the rules and write novel material each time for others to be able to publish a mosaic of their past writings as a new article.
The current incentive system rewards those who publish often. If there is no penalty for regurgitating previously published material, then we have a formula for undermining originality and creativity and honouring laziness and intellectual theft.
Excerpted from a recent publication https://www.timeshighereducation.com/opinion/we-must-take-academic-plagiarism-seriously
https://www.nytimes.com/2017/03/08/science/cancer-carlo-croce.html
Researcher hits bulls-eye for antibiotic target
Sanders worked with a team to create the first-of-its-kind animated movie showing this process from the point of view of the substrate. The audience follows along as it is pulled through the protein from one side to the next.
"This is the first time this sort of thing has ever been seen, and this is the first movie of its kind," he said. "It elegantly illustrates the physical process of this reaction." Download animation (50 MB)
ABSTRACT: Structural basis of the processivity of an exopolyphosphatase: Fusion of an ASKHA phosphotransferase and a cyclic nucleotide phosphodiesterase homologue
Johnjeff Alvarado, Anita Ghosh, Tyler Janovitz, Andrew Jauregui, Miriam S. Hasson, and David Avram Sanders
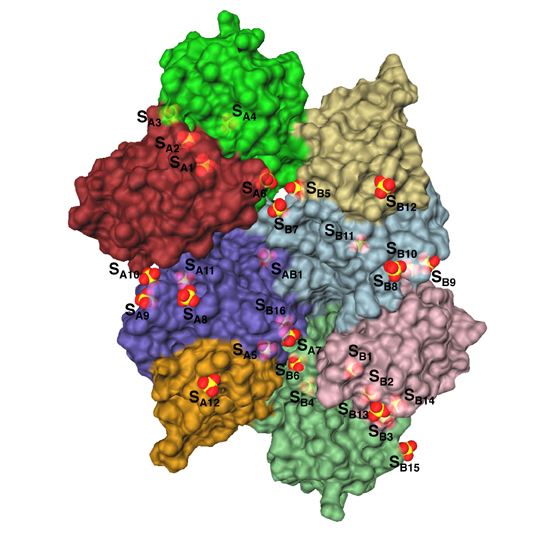
The Escherichia coli Ppx protein is an exopolyphosphatase that degrades long-chain polyphosphates in a highly processive reaction. It also hydrolyzes the terminal 5' phosphate of the modified nucleotide guanosine 5' triphoshpate 3' diphosphate (pppGpp). The structure of Ppx has been determined to 1.9 angstrom resolution by X-ray crystallography. The exopolyphospatase is an ASKHA (Acetate and Sugar Kinases, Hsp70, Actin) phosphotransferase with an active site found in a clet between the two amino-terminal domains. Analysis of the active site indicates that among ASKHA phosphotranferases of known structure Ppx is the closest to the ecto-nucleoside triphosphate diphosphohydrolases. A third domain forms a six-helix claw that is similar to the catalytic core of the eukaryotic cyclic nucleotide phosphodiesterases. Most of the twenty-nine sulfate ions bound to the Ppx dimmer occupy sites where the polyp chain likely binds. An aqueduct that passes through the enzyme provides a physical basis for the enzyme's high processivity.
Researcher hits bulls-eye for antibiotic target
WEST LAFAYETTE, Ind. — A Purdue University researcher has opened the door for possible antibiotic treatments for a variety of diseases by determining the structure of a protein that controls the starvation response of E. coli.
This research is applicable to the treatment of many diseases because that same protein is found in numerous harmful bacteria, including those that cause ulcers, leprosy, food poisoning, whooping cough, meningitis, sexually transmitted diseases, respiratory infections and stomach cancer, said David Sanders, an associate professor of biology. Sanders, who is part of the Markey Center for Structural Biology at Purdue, detailed his research in a paper published in the Aug. 16 issue of the journal Structure.
"This is an important discovery for the field of antibiotics, which was greatly in need of something new," Sanders said. "The antibiotics available today face a challenge of increasing resistance and failure. This research suggests a whole new approach to combat bacterial infections. In addition, this protein is an excellent antibiotic target because it only exists in bacteria and some plants, which means the treatment will only affect the targeted bacterial cells and will be harmless to human cells."
Sanders and his collaborator, Miriam Hasson, studied the structure of exopolyphosphatase, a protein in E. coli bacteria that functions as an enzyme and catalyzes chemical reactions within the bacteria. This enzyme provides the signal for bacteria to enter starvation mode and limit reproduction.
"With the ability to control the use of this signal, we can fool bacteria into thinking they are starving all the time, even when they are not; or we could never allow them to realize that they're starving, and that would kill them as well."
Researchers could design drugs to bind to the protein and keep it from being used by the bacteria, rendering the bacteria unable to react to and survive a lack of nutrient supply; the other possibility would be to design a drug to mimic the protein, causing the bacteria to react as if it were starving even when in the presence of a plentiful nutrient supply, Sanders said.
Such a signal exists in almost all living things because most organisms struggle to find food or nutrients and have had to develop a way to avoid starvation, he said.
"Bacteria typically are in an environment lacking nutrients and respond by limiting their reproduction," he said. "And that's a good thing because if they were growing at their maximum rate all the time, within two weeks we would be 20 feet deep in bacteria."
The protein also is of particular interest because it is highly processive, meaning it is efficient in the chemical reaction it initiates. It is able to latch onto its substrate, the substance it uses to fuel its chemical reaction, and to stay tenaciously in place until it has consumed all of the substrate, Sanders said.
Using X-ray crystallography, Sanders was able to show the structure of the E. coli exopolyphosphatase and found the protein had a unique way of achieving its high processivity.
"There is a hole in the protein," he said. "This is extremely rare and provides a physical explanation of why it is so processive. The hole physically encompasses the substrate, keeping it in place, in addition to the usual chemical bonding that keeps it attached. Once the protein attaches to the substrate, it doesn't come off. The protein chews away until it reaches the end of the substrate chain."
Sanders worked with a team to create the first-of-its-kind animated movie showing this process from the point of view of the substrate. The audience follows along as it is pulled through the protein from one side to the next.
"This is the first time this sort of thing has ever been seen, and this is the first movie of its kind," he said. "It elegantly illustrates the physical process of this reaction."
Sanders also determined the structure of the protein and demonstrated that it belongs to the ASKHA (Acetate and Sugar Kinases, Hsp70, Actin) superfamily. Knowing the family to which a protein belongs allows researchers to use existing information about other members of the family to better understand the protein being studied. It also allows information gained from the study to be used for other members of the family, Sanders said.
"Fundamental basic research is the engine that drives the development of technology such as antibiotics," he said. "The next step in this research will be working to develop inhibitors for this protein and studying the applications to other bacteria."
This research was funded by the National Institutes of Health, David and Lucille Packard Foundation Fellowship, National Science Foundation Minority Fellowship, and NIH Institutional Training Award. The NIH-sponsored Cancer Center, part of Purdue's Oncological Sciences Center in Discovery Park also supported the research.