(Return to top of document)
I. INTRODUCTION AND BACKGROUND
Agrobacterium-mediated transformation is the most commonly used method for the genetic alteration of plant cells. Scientists currently understand reasonably well the molecular events that occur in the bacterial cell prior to T-DNA transfer, including the induction of the virulence (vir) genes (103), T-DNA processing from the Ti-plasmid (29, 94), and the formation of bacterial channels for exporting the T-DNA (46,95,101), possibly as a DNA-protein complex (the T-complex; 37). However, despite the wide application of Agrobacterium to the genetic transformation of plants, we do not understand many details of the unique and complex interactions between the bacteria and plant cells. In particular, we know little about host factors involved in crown gall tumorigenesis. There are at least four steps in which plant factors participate in the Agrobacterium-mediated transformation process: Bacterial attachment to the plant cell surface, transfer of T-DNA from bacteria to plant cells across the plant wall and membrane, nuclear transport of the T-complex, and stable integration of T-DNA into the plant genome (91,108,109). These events may involve direct interactions between plant proteins and Agrobacterium virulence (Vir) proteins that are exported to the plant and accompany the T-DNA on its journey through the plant cell to the nucleus (23,91). Plant proteins involved in these infection-related events are likely also involved in basic cellular processes such as wall biosynthesis, nuclear protein and nucleic acid trafficking, and DNA repair and recombination.
(Return to top of document)
A. VirB proteins mediate T-DNA transport from Agrobacterium to the plant cell. Agrobacterium elicits neoplastic growths on many plant species. This genetic transformation is achieved by transporting a single-stranded copy (T-strand) of the bacterial transferred DNA (T-DNA) from the tumor-inducing (Ti) plasmid into the plant cell followed by integration into the host genome (reviewed in 91). Protein products of the Agrobacterium virB locus form a pilus that has been proposed as a channel through which T-DNA and its cognate proteins presumably are transferred into the host plant cell (30). From the 11 open reading frames of the virB operon (101), one protein, VirB2, encodes a pilin-like protein that resides in both the inner and outer bacterial membranes (92) and represents the major protein species found in the purified pilus preparation (48). Furthermore, VirB2 can form long extracellular filaments (48) potentially directly involved in recognition of the host cell and penetration into its cytoplasm. Besides VirB2, VirB3 and VirB9 associate with the outer bacterial membrane and, thus, may participate in interaction with the host cell.
B. VirD2 and VirE2 mediate nuclear import of T-DNA and T-DNA integration. Agrobacterium T-DNA is delimited by two 25-bp direct repeats at its ends, the T-DNA borders. The bacterial VirD2 endonuclease cleaves the bottom strand of the T-DNA borders, thereby producing the transferable T-strand (reviewed in 91). While only the wild-type T-DNA carries tumor-inducing genes, any DNA placed between the T-DNA borders will be transported into the plant cell nucleus (reviewed in 105). This lack of sequence specificity implies that a T-DNA molecule itself does not contain specific signals for nuclear import. Instead, this process is likely mediated by two Agrobacterium proteins, VirD2 and VirE2, that are thought to associate directly with the T-strand, forming a transport (T) complex (reviewed in 91). In the T-complex, one molecule of VirD2 becomes covalently attached to the 5' end of the T-strand following cleavage of the T-DNA borders while VirE2, a single-stranded (ss) DNA binding protein (15,19,33,88), is presumed to coat the rest of the ssDNA molecule (reviewed in 14), shaping it into a coiled telephone cord-like structure (16). Our studies showed that VirD2 contains one functional bipartite nuclear localization signal (NLS) and VirE2 contains two bipartite NLSs (18,20,38,43,44,63).
To specify the polarity of the T-complex and/or to prevent competition between the single NLS of VirD2 and the NLSs of numerous VirE2 molecules, these signals may interact with different NLS receptors. Indeed, our recent results indicate that the nopaline-type VirE2 NLSs may be plant-specific (35) while the NLS of VirD2 functions both in plant and animal cells (35,77). The role of VirE2 in T-complex nuclear uptake was confirmed directly using microinjection of fluorescently-labeled VirE2-ssDNA complexes into stamen hair cells of Tradescantia virginiana. While fluorescent ssDNA remained in the cell cytoplasm when microinjected alone, it accumulated in the cell nucleus when microinjected in complexes with VirE2 (107).
Nuclear import of the T-complex culminates with T-DNA integration into the host genome. The mechanism by which T-DNA integration occurs is largely unknown. Unlike other mobile DNA elements such as transposons and retroviruses, T-DNA does not encode enzymatic activities required for integration. Thus, T-DNA insertion into the plant DNA must be mediated by proteins transported from Agrobacterium itself and/or host cell factors. Indeed, both VirD2 and VirE2 proteins of the T-complex have been implicated in the integration process (63,67,72,83,97). Specifically, an amino acid sequence downstream of VirD2 NLS, the omega domain (93), is likely involved in T-DNA integration (63,67). Also, an R to G mutation in the H-R-Y integrase motif of VirD2 decreases the precision but not the efficiency of T-DNA integration in vivo (96). Finally, to complete integration, VirD2 may participate in the ligation of the 5' end of the T-DNA to the genomic DNA in vivo (97). VirE2, on the other hand, is required for integration fidelity of the T-DNA 3' end (83).
(Return to top of document)
C. VirF functions in planta and expands the host range of Agrobacterium. The presence of a virF locus expands the host range of several Agrobacterium species, e.g. A. tumefaciens and A. vitis (87 and references therein). Interestingly, expression of VirF in transgenic non-host Nicotiana glauca plants makes them susceptible to Agrobacterium (75). In addition, coinoculation experiments demonstrated that VirF is likely transported from Agrobacterium to plant cells during the infection process (71). While these results indicate that the VirF protein may perform an important function within the plant, its role in the infection process is still unknown.
Considering the importance of Vir proteins in the Agrobacterium transformation process, we have targeted for our research plant factors that are likely to interact with these bacterial proteins.
II. PRELIMINARY RESULTS
A. Isolation of Arabidopsis mutants that are resistant to Agrobacterium transformation. One of us (SBG) has recently initiated a project to identify and characterize Arabidopsis thaliana mutants that are resistant to transformation by Agrobacterium tumefaciens (65). In summary, we have screened about 3,000 individual M4 generation T-DNA mutagenized plants from the Feldmann T-DNA insertion collection (26,27) and have identified 21 rat (resistant to Agrobacterium transformation) mutants among these lines. These mutants were tested for transformability by assaying crown gall tumorigenesis, transformation to phosphinothricin (ppt)-resistance, and transformation to express GUS activity both transiently and stably (Table 1).
Table 1. Characterization of Arabidopsis rat mutants
| Line |
Transient Transformationa
% Root Bundles Showing
GUS Staining |
Stable Transformationb
% Root Bundles Showing
ppt-Resistance Tumorigenicity |
Probable Function of RAT Genec |
Wild-type |
92± 6 |
87± 10 |
86± 15 |
- |
| rat1 |
22± 4 |
5± 2 |
7± 1 |
arabinogalactan protein |
| rat3 |
31± 2 |
9± 2 |
10± 4 |
likely cell wall protein |
| rat4 |
10± 4 |
14± 4 |
19± 8 |
cellulose synthase-like protein |
| rat5 |
86± 2 |
15± 5 |
8± 3 |
histone H2A |
| rat6 |
9± 2 |
12± 10 |
16± 5 |
not sequenced |
| rat7 |
18± 10 |
10± 5 |
18± 10 |
unknown |
| rat8 |
30± 8 |
40± 5 |
36± 5 |
not sequenced |
| rat9 |
20± 4 |
6± 4 |
4± 5 |
unknown |
| rat10 |
26± 12 |
19± 5 |
3± 4 |
not sequenced |
| rat11 |
24± 4 |
14± 3 |
4± 5 |
not sequenced |
| rat12 |
19± 5 |
10± 5 |
5± 6 |
not sequenced |
| rat13 |
11± 2 |
20± 10 |
5± 8 |
not sequenced |
| rat14 |
13± 1 |
9± 5 |
6± 5 |
unknown |
| rat15 |
18± 2 |
11± 6 |
13± 5 |
not sequenced |
| rat16 |
28± 7 |
33± 11 |
21± 10 |
not sequenced |
| rat17 |
90± 6 |
20± 12 |
18± 12 |
myb-like transcription factor |
| rat18 |
88± 5 |
27± 8 |
21± 7 |
not sequenced |
| rat19 |
18± 3 |
9± 4 |
4± 3 |
not sequenced |
| rat20 |
83± 10 |
31± 10 |
10± 6 |
not sequenced |
| rat21 |
20± 8 |
21± 8 |
8± 9 |
not sequenced |
| rat22 |
70± 5 |
29± 15 |
17± 8 |
unknown |
a 3 plants for each mutant and >100 root segments for each plant were examined; b 5 plants were tested for each mutant and 40 - 50 root bundles were tested for each plant; c the proposed gene function is based on sequence homology of cDNA and genomic clones identified by the T-DNA-plant DNA junctions. For further detail, see (65).
In all but one of the isolated mutants, the rat phenotype co-segregated with the kanamycin-resistance marker on the mutagenic T-DNA. Sixteen of these rat mutants (rat1, 3, 4, 6-16, 19, 21) appeared to be blocked at an early stage of the transformation process because their infection by an Agrobacterium strain carrying T-DNA with a reporter gusA-intron gene resulted in very low levels of transient GUS expression (Table 1), indicating deficiencies in T-DNA transport into the plant cell and/or into its nucleus. In contrast, five of the mutants (rat5, 17, 18, 20, 22) are probably blocked at a late (likely T-DNA integration) step in the transformation process because their infection resulted in a high degree of transient GUS expression but low levels of stable transformation (Table 1).
To date, we have recovered T-DNA/plant DNA junctions from 9 rat mutants and analyzed the sequence of the plant DNA (Table 1). At least three of the disrupted genes likely play an important role in plant cell wall structure and biosynthesis. The disrupted RAT1 gene encodes an arabinogalactan protein (AGP). We are aware of only one other report of an AGP mutant, reb1 (24). Agrobacterium cells do not attach to roots of rat1 plants either in water or in medium containing sucrose. Thus, the rat1 mutant may lack a plant cell surface component necessary for Agrobacterium to bind. AGP involvement in Agrobacterium infection is supported by our observations that the beta-glucosyl Yariv reagent, which binds to AGPs, inhibited transformation of Arabidopsis root segments by Agrobacterium, whereas the control beta-mannosyl Yariv reagent did not.
The gene disrupted in the rat4 mutant encodes a cellulose synthase-like (Csl) protein (Table 1). This is, as far as we are aware, the only csl mutant plant identified to date. Csl proteins function as synthases of non-cellulosic polysaccharides (T. Richmond, P. Villand, S. Cutler, and C. Somerville, poster presented at the 9th International Conference on Arabidopsis Research, Madison, WI, 1998). In Arabidopsis, 14 CSL genes were identified by their homology to the cellulose synthase (CELA) multigene family that in turn comprises at least 30 genes (1).
The rat3 mutation contains a T-DNA insertion in a 600 bp area between two highly homologous genes. The 15 kDa proteins encoded by this two-gene family do not show homology to any genes in the data bases. The proteins have a signal peptide, and also contain a GPI anchor domain in their C-termini. Thus, these proteins are likely targeted to the cell surface. rat3 is deficient in Agrobacterium binding in water, although this deficiency is not so pronounced in medium containing sucrose.
Among the putative T-DNA integration-deficient mutants are rat5 (a histone H2A mutant) and rat17 (a myb-like transcription factor mutant) (Table 1). RAT5 is one of a five-member histone H2A gene family in Arabidopsis. We have complemented the rat5 mutant with the corresponding histone H2A gene, and are currently conducting complementation tests with rat17.
(Return to top of document)
B. NLS signals of VirD2 and VirE2 proteins are functionally different. Numerous VirE2 molecules associated with the T-strand likely supply a large number of NLSs (16). Still, the single NLS of VirD2 appears to play a role in the T-complex nuclear import because its specific deletion often reduced tumorigenesis and T-strand nuclear accumulation (67,96,104). We hypothesized that VirE2 and VirD2 NLSs may be functionally distinct, interacting with different receptors within the host plant cell. These differences became apparent in a heterologous animal system after we microinjected fluorescently-tagged nopaline-type VirD2 and VirE2 proteins into 1.5-2 hour-old Drosophila embryos that contained ~6000 nuclei in a syncytium (12).
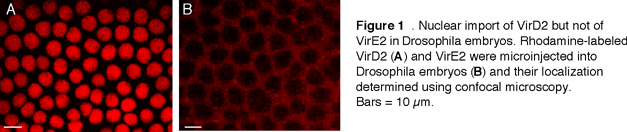
Microinjected VirD2 accumulated in Drosophila nuclei (Figure 1A); this nuclear uptake was blocked by free NLS peptide as well as by coinjection of a known nuclear import inhibitor GTPgS (data not shown but see 35). [Note that our microscopic data are confocal optical sections with the plane of focus through the cell nuclei.]. Unlike VirD2, nopaline-type VirE2 remained cytoplasmic in Drosophila embryos (Figure 1B), Xenopus oocytes (35), yeast, and HeLa cells (data not shown). This is in contrast to the efficient nuclear import of VirE2 in different plant species (18). Thus, the complete absence of VirE2 nuclear import in animal cells indicates that the VirE2 NLS may be plant-specific. Interestingly, a single amino acid change in the VirE2 NLS redirected this protein into the cell nuclei of Drosophila embryos and Xenopus oocytes (35).
C. Identification of AtKAP(alpha), an Arabidopsis protein that recognizes the VirD2 NLS. We began our study of plant factors that interact with the invading T-complex with identification of an Arabidopsis protein that binds the VirD2 NLS. Using a yeast two-hybrid screen (28) with VirD2 as bait, we identified an Arabidopsis cDNA clone, designated AtKAP(alpha) (Arabidopsis thaliana karyopherin(alpha), after eliminating the false positives using a non-specific lamin C bait (5) and a VirD2 mutant lacking the NLS (VirD2(delta)NLS, Figure 2). We isolated the full length cDNA of AtKAP(alpha) and confirmed its specific interaction with VirD2 by coimmunoprecipitation (4).
AtKAP(alpha) cDNA contained a single ORF encoding a protein of 65.6 kDa with 44% identity to yeast Srp1 (4), a member of the karyopherin(alpha) family of proteins that function as NLS receptors (47,53,102). Indeed, AtKAP(alpha) complemented the srp1-31 yeast mutant (4,53) and was shown directly to transport fluorescently-labeled VirD2 into the nuclei of permeabilized yeast cells (4).

We suggest that, during Agrobacterium infection, AtKAP(alpha) recognizes VirD2 and promotes its nuclear import. Because VirD2 is tightly associated with the T-strand (reviewed in 91) AtKAP(alpha) likely mediates nuclear import of the T-complex as well. Unlike VirD2, VirE2 did not interact with AtKAP(alpha) in the two-hybrid assay (data not shown). Thus, another as yet unidentified plant protein that recognizes VirE2 NLSs may also be involved in T-complex nuclear import. In uninfected plants, AtKA(alpha) may function as a NLS-binding protein mediating transport of cellular nuclear proteins.
(Return to top of document)
D. A type 2C protein phosphatase potentiates nuclear import of VirD2 protein. We used a yeast two-hybrid system to identify additional host proteins that interact specifically with the NLS region of VirD2 (Tao and Gelvin, in preparation). From a tomato two-hybrid cDNA library, one clone, DIG3, interacted with a VirD2 bait containing the C-terminal bipartite NLS, but did not interact with a bait containing a precise deletion of this signal. DIG3 encodes a Type 2C protein phosphatase (PP2C); we have demonstrated PP2C enzymatic activity of a GST-DIG3 fusion protein. Because the Arabidopsis abi1 mutant (a PP2C mutant) showed increased susceptibility to transformation by Agrobacterium, we suggested that a PP2C is a negative regulator of Agrobacterium transformation.
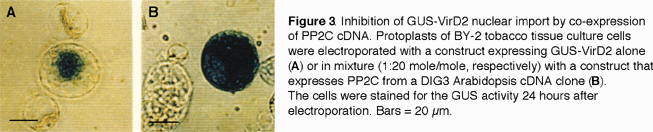
When we electroporated a GUS-VirD2 NLS fusion gene into tobacco BY-2 protoplasts, GUS activity localized exclusively to the nucleus in approximately 80% of the cells (Figure 3A). Co-electroporation of the PP2C gene sharply reduced GUS nuclear localization (Figure 3B), indicating that PP2C indeed negatively regulates nuclear trafficking of VirD2. We identified a consensus protein kinase C phosphorylation site on a serine residue (Ser-424) two amino acids upstream of the VirD2 NLS. We detected phosphorylation of Ser-424 both in vitro following incubation with protein kinase C and in vivo in tobacco protoplasts. Alteration of Ser-424 to alanine resulted in decreased nuclear import of a GUS-VirD2 mutant NLS fusion protein in tobacco protoplasts, and decreased tumorigenesis of an Agrobacterium strain containing this mutant VirD2 protein. We thus propose that phosphorylation of the serine residue near the VirD2 NLS potentiates T-complex nuclear import, and that the activity of a plant PP2C decreases VirD2 nuclear import and, hence, transformation by Agrobacterium (Tao and Gelvin, in preparation).
E. Development of in vitro T-DNA integration assays. In higher eukaryotic organisms such as plants, DNA integration occurs predominantly by illegitimate recombination, i.e., two DNA molecules that do not share extensive homology are joined (60,69,73,85). Illegitimate recombination is also responsible for T-DNA integration into the plant genome during Agrobacterium infection (31,56,57,70,97). The role of bacterial and/or plant factors in this process is still unclear. VirD2 has been shown to cleave the T-DNA border sequence of ssDNA efficiently in vitro as well as to ligate the adjoining DNA strands (40,72). Although VirD2-mediated in vitro cleavage and rejoining reactions are T-DNA border specific, T-DNA integration in vivo is sequence independent (31,56,57,97). To understand better the involvement of host cell factors in the integration process, we (BH) have designed an in vitro assay for ligation of the 5' end of T-DNA to the plant DNA. In this approach, a VirD2-T-DNA complex was formed in vitro and ligated to an artificial target DNA (Figure 4) The VirD2-T-DNA complex was produced by VirD2-dependent cleavage of a 35-mer oligonucleotide containing the border sequence. During this reaction, VirD2 becomes covalently attached to the 5' end of an 8-mer cleavage product via a phosphotyrosine bond (72). The artificial target DNA contained single-stranded and double-stranded regions. The single-stranded part was homologous to the 5' end of the T-DNA to facilitate annealing whereas the double-stranded part was not homologous to the T-DNA, mimicking plant genomic DNA. Integration of the VirD2-T-DNA complex into the target DNA was monitored by the appearance of a radiolabeled 21-mer oligonucleotide (Figure 4).
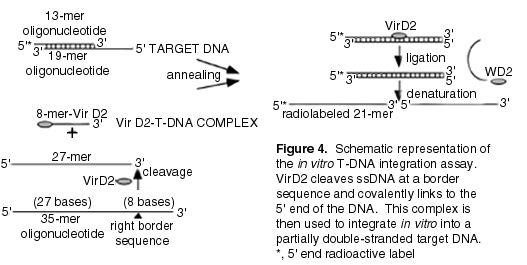
In this in vitro system, the sequence-specific integration of the VirD2-T-DNA complex occurred with high efficiency (data not shown but see ref. 106). This is consistent with the previously reported activity of VirD2 in rejoining oligonucleotides containing complementary parts of the border sequence (72). However, VirD2 was unable to mediate ligation to a non-specific target DNA suggesting that plant cell ligases and possibly other factors are required for T-DNA integration. Because a purified plant DNA ligase is not yet available, plant extracts were tested for their ability to ligate T-DNA to plant DNA in vitro. We could detect low activity in extracts from tobacco BY-2 cells and pea shoot apices (data not shown).
(Return to top of document)
III. RELEVANCE
Agrobacterium-mediated transformation is currently the major technology for the genetic manipulation of plants, both for research and for agricultural biotechnology purposes. Although a large number of agronomic and horticultural plants are susceptible to Agrobacterium-mediated genetic transformation, many important species (or elite cultivars/lines of species) are recalcitrant. Thus, the efficient transformation and regeneration of crop plants remain a priority for the plant science and biotechnology community. Although researchers have developed several "improved" strains of Agrobacterium that are better able to transform plants, we have probably neared the limit in improving crop transformation by manipulation of the bacterium. Future improvements will necessarily come from a better understanding, and the subsequent manipulation, of the plant host. Unfortunately, while scientists currently have a reasonably complete understanding of the molecular and genetic events that occur within Agrobacterium during transformation, little is known about the role of specific plant genes and proteins in this process. We do know that closely related cultivars or ecotypes of specific plant species can differ greatly in transformation-proficiency, and we have recently been able to isolate a large number of Arabidopsis mutants that have lost susceptibility to Agrobacterium transformation. The identification of plant genes and proteins necessary for transformation, and the manipulation of these genes in recalcitrant crop species, is therefore the next route for improving plant transformation technology.
This proposal represents a comprehensive approach to understanding the role of host genes and proteins in Agrobacterium-mediated transformation. We have brought together three laboratories with great expertise in investigating Agrobacterium-plant interactions from the plant's perspective. These groups collectively will conduct extensive genetic, molecular, and biochemical analyses to identify plant genes involved in Agrobacterium transformation. We expect that the fruits of our endeavors will benefit the plant community in four essential ways: (i) By identifying plant genes essential for transformation, we shall be able to manipulate these genes to effect a more efficient transformation of currently-recalcitrant species; (ii) By identifying plant genes essential for transformation, we shall be able to create crown gall-resistant plants by the genetic manipulation of these genes; (iii) We shall increase the efficiency of gene targeting, relative to random integration, by utilizing plants that are deficient in integrating T-DNA by illegitimate recombination (this specific goal is the subject of a separate grant to SBG and thus it is not included in our proposal); (iv) Because plant genes necessary for Agrobacterium transformation are involved in plant cell processes such as wall biosynthesis and structure, macromolecular transport (including nuclear import), and DNA repair and recombination, our research will result in a greater understanding of basic plant cell biology.
(Return to top of document)
IV. SPECIFIC AIMS
A. Identification and characterization of Arabidopsis rat mutants and molecular cloning of RAT genes. We shall expand our genetic screen for rat mutants with the final goal to examine 200,000 unique T-DNA insertion mutations that saturate the entire Arabidopsis genome. We shall characterize the isolated rat mutants genetically and biochemically to identify the specific stage at which the transformation process is arrested. The T-DNA tagged RAT genes will then be cloned and tested for their ability to complement the corresponding rat mutants.
B. Identification of Arabidopsis proteins that directly interact with Agrobacterium Vir proteins. Presently, four Agrobacterium proteins are known to interact directly with host cell components during the infection process. VirF likely functions in planta, determining the host range of Agrobacterium; VirB2 acts as a pilin that mediates transfer of DNA from the bacterium into the host plant cell; VirD2 and VirE2 presumably associate with the T-DNA molecule, transport it into the host cell nucleus, and assist in its integration into the plant genome. We shall utilize the yeast two- and one-hybrid systems to identify plant proteins that directly interact with VirF, VirB2, VirD2 and VirE2 and are involved in Agrobacterium-plant cell recognition, intercellular transport of T-DNA, and its nuclear import and integration.
C. Identification of Arabidopsis genes induced or repressed during the initial stages of Agrobacterium transformation. Agrobacterium infection represents a major physiological, biochemical, and genetic challenge to the host plant. Most likely, this event triggers changes in host cell gene expression patterns. In this proposal, we shall identify plant genes specifically induced or repressed during the early stages of the Agrobacterium infection process using differential display, microarray, and subtraction library approaches. We shall compare gene expression in healthy Arabidopsis plants and cell cultures to that in plants exposed to Agrobacterium.
D. Biological characterization of Arabidopsis genes involved in Agrobacterium-mediated transformation. We shall examine the biological activities of the identified host genes and proteins using specific assays developed in each of the collaborating laboratories. For example, we shall use reverse genetics to identify and phenotypically characterize Arabidopsis mutants in genes isolated in two- and one-hybrid screens, differential display, subtraction, and microarray analyses. We shall analyze VirD2- and VirE2-interacting proteins for their ability to promote nuclear import of VirD2, VirE2, and VirE2-ssDNA complexes in animal, yeast and plant cells. Finally, we shall use in vitro assays to test whether VirD2- and VirE2-interactors are involved in T-DNA integration in vitro.
E. Improvement of agronomically important crops using Arabidopsis genes essential for Agrobacterium infection. Arabidopsis genes encoding protein factors that participate in Agrobacterium-host plant interactions will be utilized to improve the transformation of agriculturally important but currently recalcitrant maize species and to prevent crown gall disease in grape.
(Return to top of document)
V. EXPERIMENTAL PLAN
Experiments will be carried out in five stages that correspond to the specific aims of this proposal.
A. Identification and characterization of Arabidopsis rat mutants and cloning of RAT genes
1. Use of existing Arabidopsis T-DNA insertion collections
a. The Feldmann Collection. One of us (SBG) has initiated a search for Arabidopsis rat (resistant to Agrobacterium transformation) mutants (see Preliminary Results). Based on the high incidence of rat mutants found in the T-DNA tagged Feldmann collection (0.7% of the approximately 3,000 mutant lines tested to date), and the number of genes in Arabidopsis (20,000-30,000), we estimate that a total of 200 to 300 Arabidopsis genes may be involved in the rat phenotype (65). This large number of genes is not without precedent. For example, several laboratories have reported a high frequency of Arabidopsis mutants hypersensitive to radiation (22,55). Given the complexity of the Agrobacterium infection (e.g., bacterial binding to the plant cell wall and/or cell membrane and T-DNA transfer, nuclear import, and integration into the plant genome), the large number of plant genes involved in this process makes biological sense. Some of these genes directly participate in the transformation process. For example, RAT1 encodes a cell wall arabinogalactan protein that is required for Agrobacterium binding to the root surface (65). RAT5 encodes a histone H2A variant potentially involved in T-DNA integration (Mysore et al., in preparation). Other RAT genes may only indirectly affect Agrobacterium infection. For example, RAT17 encodes a myb-like transcription factor implicated in root development (100). The rat17 mutant is another allele of the caprice mutant. Possibly, disruption of this gene renders plant roots recalcitrant to Agrobacterium transformation.
Our initial screening of 3,000 mutagenized plants is not a comprehensive analysis of the existing collections of Arabidopsis T-DNA mutants. First, because the Feldmann collection represents pools of 100 plants, some of the analyzed plants may have been sibs, effectively reducing the number of independent mutants screened. Second, this collection contains about 6,400 individual mutagenized plants, with an average 1.5 independent T-DNA insertions per plant (2), corresponding to approximately 10,000 separate insertion events. Thus, the Feldmann collection does not tag all of the 20,000 to 30,000 Arabidopsis genes.
b. Use of Other Collections to Achieve Saturation Mutagenesis. Comprehensive analysis of the host genes involved in Agrobacterium transformation requires saturation mutagenesis of the Arabidopsis genome. To expand the mutant pool, therefore, we have (or shall soon) obtained three additional collections of T-DNA mutagenized Arabidopsis plants. The DuPont collection, comprising approximately 7,200 individual members with an average of 1.5 independent T-DNA insertions per plant, is expected to tag 11,000 genes (Tim Caspar, personal communication). The Amasino/Sussman ArabidopsisT-DNA collection (see attached letter) currently includes approximately 70,000 T3 generation T-DNA "disruption-tagged" lines (that are currently being released for distribution by The Ohio State University Arabidopsis Stock Center) and will soon include another 70,000 T-DNA "activation-tagged" lines (Rick Amasino, personal communication). Finally, as part of an NSF-funded Plant Genomics Project, Dr. Ray Bressan (Department of Horticulture, Purdue University) is establishing a collection of T-DNA "activation-tagged" Arabidopsis lines that will be available to us as they are developed (see attached letter).
Collectively, these T-DNA-tagged Arabidopsis collections represent more than 200,000 unique T-DNA insertion events and should saturate the Arabidopsis genome with tagged mutations.
2. rat mutant screening procedures
We plan to continue screening the T-DNA-tagged Arabidopsis collections for rat mutants as described (65). Based on our experience, one person at full-time effort can easily screen between 3,000 to 4,000 plants per month. This throughput will allow two technicians and several undergraduate students (at Purdue) and one postdoc (at Stony Brook) requested in the proposed budget to screen 200,000 independent mutants from all the available T-DNA insertion collections within a two year period. As mentioned above, this approach identifies rat mutants at the frequency of 0.7% of the screened population. Thus, we are certain that additional screening experiments will identify many new mutants with interesting phenotypes.
Technically, the screening procedure involves sterilizing seeds, plating them on a sterile solidified growth medium, and growing the plants for 2-3 weeks to allow development of the roots. The roots are then cut and infected with a teratoma-inducing strain of Agrobacterium, A208, that reproducibly transforms a very high percentage of Arabidopsis root segments (64). In the mean time, a shoot from each plant is replanted into sterile solidified medium for re-rooting. The shoots derived from plants whose roots did not develop teratomas are transferred to soil. The progeny of these plants are tested again for transformation by Agrobacterium, this time using a disarmed nopaline-type strain, GV3101(pCAS1) that efficiently transforms Arabidopsis, conferring phosphinothricin (ppt)-resistance upon the infected plant. In addition, the mutants are re-tested for their susceptibility to tumorigenesis following infection with the A208 strain. Because we can obtain 50-100 sterile root segments from each plant, we can perform statistical analysis to determine the percentage of roots transformed to these two stable phenotypes (i.e., teratomas and ppt-resistance). While 90% of the wild-type Arabidopsis root segments are typically transformed (65), the resistant plants should exhibit significantly lower transformation efficiencies. In this approach, plants whose root segments yield less than 10% transformation will be identified as rat mutants and subjected to further analyses.
3. Which stage of the transformation process is blocked in the rat mutants?
Initially, we shall determine whether the identified mutants are blocked at an early or late stage of the Agrobacterium transformation process. To this end, we shall infect sterile root segments of these mutants with A. tumefaciens GV3101 containing the T-DNA binary vector pBISN1. pBISN1 (67) carries within its T-DNA a nos-nptII-polyA selectable marker and a gusA-intron gene under the transcriptional control of a "super-promoter" (68). This promoter is extremely active in Arabidopsis (64), allowing us to detect GUS expression 2 days after infection. Importantly, this assay specifically detects GUS activity within the transformed cells because the intron in the gusA gene prevents GUS expression in Agrobacterium (52). We shall assay for two types of GUS expression: transient and stable. For transient GUS expression, we shall stain the root segments of the rat mutants and the wild-type parental ecotype with X-gluc at 2 and 5 days after infection. Positive staining indicates that T-DNA has been transferred to the plant, has targeted to the nucleus, and has been converted to a double-stranded form required for transcription. However, transient GUS expression does not necessarily indicate that the T-DNA has integrated into the Arabidopsis genome. We shall continue to grow the root segments for 30 to 40 days on medium containing antibiotics to eliminate Agrobacterium, and phytohormones to obtain calli which will then be assayed for stable GUS activity. Note that we are not selecting for tumorigenesis or plant antibiotic or herbicide resistance. We have previously demonstrated that, in such unselected calli, transient GUS activity declines to undetectable levels by 30 to 40 days after infection, whereas stable GUS expression from the integrated T-DNA can still be detected (64). Thus, Arabidopsis plants with high levels of transient GUS activity but low or no stable GUS expression will likely represent integration-deficient rat mutants. In contrast, mutants with negligible or low levels of transient GUS expression will represent rat mutations that block early stages of the Agrobacterium infection.
a. Integration-Deficient rat Mutants. We shall confirm that these rat mutants are T-DNA integration deficient using two experimental approaches. First, we shall infect root segments with Agrobacterium GV3101 that carries the promoter-trap T-DNA binary vector pKM1 (63) containing a promoterless gusA-intron gene with its 5' end in close proximity to the T-DNA right border. Because this reporter gene expresses only after integration downstream of an active plant promoter, it can be used as a functional marker for an integration event (63). To control for T-DNA transfer and nuclear import that precede integration, pKM1 contains a luc-intron (luciferase) gene under the control of the 35S promoter that is expressed regardless of T-DNA integration. Plants with transient luciferase activity but no GUS staining will be classified as putative T-DNA integration-deficient mutants.
Second, we shall generate cell culture lines from the rat mutants that appear to be T-DNA integration-deficient and assay these cells directly for T-DNA integration. To this end, we shall infect Arabidopsis cell cultures for 2 days with Agrobacterium GV3101 containing pBISN1 carrying the gusA-intron gene for 2 days, followed by killing the bacteria with antibiotics and continuation of cell growth with no selection for any transformation phenotype for an additional 4-6 weeks. We shall isolate high molecular weight plant DNA, subject it to electrophoresis though a 0.7% agarose gel (without restriction digestion), blot the DNA onto a membrane, and hybridize the blot with a gusA-specific probe. A positive hybridization signal will directly indicate T-DNA integration into rat mutant high molecular weight DNA. To assure that we are not detecting hybridization to DNA from contaminating bacteria, we shall rehybridize the blot with an Agrobacterium chromosomal gene (picA; 80, 81); lack of hybridization would indicate the absence of detectable contaminating bacteria. Finally, we shall rehybridize the blot with an Arabidopsis phenylalanine ammonia lyase (PAL) gene to assure equal loading of the lanes (64). We shall, of course, be looking for rat mutants that do not contain T-DNA integrated into the plant genome. As a positive control for integration, we shall conduct the same experiments using wild-type cell suspensions. As a negative control, we shall use an Agrobacterium strain (A136) that contains pBISN1 but lacks a Ti-plasmid (which provides Vir protein functions for T-DNA transfer). We have previously utilized this assay to show that a particular Arabidopsis ecotype (UE-1) is deficient in T-DNA integration (64) and that a virD2 omega-mutant Agrobacterium strain is deficient in its ability to integrate T-DNA into the wild-type Arabidopsis genome (63). We shall compare our in vivo and in vitro T-DNA integration results (see below).
b. rat Mutants Blocked at Early Stages of Interaction with Agrobacterium. Mutants that show neither transient nor stable GUS expression (nor transformation to the other stable phenotypes of teratomas and ppt-resistance) are blocked at an early stage of the transformation process. We shall first examine these mutants for their ability to bind Agrobacterium cells. Briefly, we shall incubate sterile root segments for 24 hours with various concentrations of Agrobacterium cells in either water or sucrose. The root segments are then gently washed and examined using a microscope with Nomarski optics (64,65). rat plants that still bind bacteria but are resistant to transient (and stable) transformation will be classified as mutants with a block in T-DNA transfer or nuclear import.
4. Genetic characterization of Arabidopsis rat mutants
a. Identification of Homozygous rat Plants. We shall perform genetic analyses to characterize our mutants further. Generally, we would like to conduct all further analyses on homozygous plants. However, many of the mutants in the Feldmann and DuPont collections are heterozygous for the T-DNA insertion. Thus, we shall first segregate the mutagenic T-DNA to produce homozygous rat plants. To this end, we shall allow the plants to self-fertilize and test roots of the progeny for kanamycin-resistant callus formation. If the plants are homozygous, all progeny should be kanamycin-resistant, i.e. carrying the T-DNA insert. If the mutants segregate kanamycin-sensitive plants, this indicates that they are heterozygous. In this case, we shall test the progeny of the self-pollinated kanamycin-resistant plants to identify homozygous plants.
There may be two situations that would complicate our genetic analyses. We may not be able to generate plants that yield only kanamycin-resistant progeny. Especially if the progeny segregate 2:1 kanamycin-resistant:kanamycin-sensitive, this would indicate that the mutation in the homozygous condition is lethal and that kanamycin resistance reflects the dominant phenotype of this gene. We should be able to work with the heterozygote for further analysis, but the genetics would be slightly more complicated than working with the homozygote. Furthermore, it is also possible that even if only kanamycin-resistant progeny are recovered, the plant is not homozygous for one mutagenic T-DNA insertion, but rather that multiple T-DNAs inserted into several independent linkage groups, marking them with the kanamycin-resistance gene. It is likely that only one of these insertions would be in a rat locus. This would make the mutant very difficult to analyze. Such a situation would be uncovered in progeny analysis of crosses of the mutant plant with the wild-type plant. F2 progeny would not segregate 3:1 kanamycin-resistant:kanamycin-sensitive (because kanamycin resistance is a dominant phenotype, they would segregate 15:1 if there were 2 independently segregating mutagenized loci). It may be possible to segregate the two T-DNA inserts from each other and subsequently test for co-segregation of the kanamycin-resistance with the mutant phenotype, but this would be very difficult to do if there were more than two independently segregating mutagenized loci. If such were the situation, we would most likely abandon analysis of the particular mutant.
b. Dominance/Recessivity and Co-Segregation Tests. Next, we shall cross each of the kanamycin-resistant rat mutant plant (pollen donor) with the kanamycin-sensitive wild-type plant (egg donor). We shall select kanamycin-resistant F1 plants and determine whether the rat phenotype is dominant, semi-dominant, or recessive. In addition, we shall subject F2 progeny of self-pollinated F1 plants to X1 analysis to determine whether the rat phenotype results from a single mutation.
Finally, we shall analyze the F2 progeny to determine whether the mutagenic T-DNA (represented by kanamycin-resistance) always co-segregates with the mutant phenotype. This analysis is essential because T-DNA insertions frequently do not co-segregate with the mutant phenotype (25,26,42,54,99). In our experience, however, the great majority of rat mutants exhibit co-segregation of the kanamycin resistance and rat phenotypes. Of course, co-segregation should not be taken as a proof that the T-DNA has tagged the gene of interest; rather, merely that the T-DNA has inserted at a position either at or very closely linked to the gene of interest. Ultimately, we shall use genetic complementation of the mutant with the wild-type gene to prove that we have correctly identified the gene involved in the mutant phenotype (see below).
5. Molecular cloning of RAT genes
a. Isolation of T-DNA/Plant DNA Junctions. Once we have established that the T-DNA has tagged or closely linked to the gene responsible for the mutant phenotype, we shall isolate the T-DNA/plant DNA junction. Ken Feldmann designed the T-DNA mutagenizing vector such that T-DNA/plant DNA junctions could be recovered by plasmid rescue (i.e., restriction endonuclease digestion of the mutant plant genomic DNA followed by self-ligation and transformation into a recA-, mcrABC- strain of E. coli). However, in our experience, junction recovery is not as straight-forward as it may seem. This is because most mutants contain multiple T-DNA insertions within a single locus. These insertions can be dimers (tandem or head-to-head) or higher multimers. Thus, there may not be any right (or left) T-DNA/plant DNA junctions. In addition, we have frequently seen bands that, on a genomic Southern blot, would appear to be junctions (i.e., not of the expected size for a tandem or head-to-head T-DNA multimer) but upon recovery and hybridization to wild-type Arabidopsis DNA show no signal. These bands are merely rearranged T-DNAs. We are well aware of these difficulties and have been able to "work our way through them". Before initiating a junction recovery by plasmid rescue, we always conduct an extensive series of genomic Southern blot analyses to determine the number of T-DNA insertions and the organization of the inserted T-DNA. Although many insertions are complex, we have been able to isolate junction clones using plasmid rescue from all rat mutants with which we have worked. If plasmid rescue fails, we shall use inverse PCR, with one primer just inside the T-DNA border and another within the T-DNA, to rescue junction fragments.
b. Isolation of RAT cDNA and Genomic Clones. Once we have obtained the putative junction clone, we shall use it as a probe in Southern blot analysis of genomic DNA from wild-type and mutant plants. If we have isolated a T-DNA junction, we should detect a RFLP between these DNA samples.
Next, we shall determine whether any DNA rearrangements have occurred during isolation of the T-DNA junction (in our experience, such rearrangements are common) by comparing the restriction maps of the junction clone and genomic DNA from the mutant. We shall then sequence the junction clone to determine the insertion position of the T-DNA within the plant DNA. We have, so far, always been able to identify the T-DNA/plant DNA junction region. Using the junction clone as a hybridization probe, we shall screen both cDNA and genomic DNA libraries of wild-type Arabidopsis. In our experience, for all mutants except one, we have been able to identify both cDNA and genomic clones corresponding to the T-DNA/plant DNA junction region. The one exception was a mutant for which we could not directly isolate a cDNA. We later discovered that this was because the T-DNA inserted rather far from the structural gene and the junction clone did not contain the gene sequence. However, we were eventually able to identify the correct cDNA using the genomic DNA clone as a hybridization probe. Another problem with which we have dealt in the past was cloning a member of a multigene family. Specifically, the rat5 mutant represented a T-DNA insertion into a histone H2A gene (Mysore et al., in preparation). Because H2A belongs to a large multigene family, the junction clone hybridized to many different genomic and cDNA clones. To isolate the clone corresponding to the particular H2A gene disrupted by T-DNA insertion, we used a unique plant DNA region in the junction clone upstream of the histone gene as a hybridization probe.
Following isolation of rat genomic DNA clones, we shall compare their restriction maps to those of the mutant genomic DNA to assure that no large deletions or rearrangements of the genome occurred during the T-DNA insertion process. A deletion could indicate that the gene of interest was not mutated by the inserted T-DNA, but was rather deleted as a consequence of T-DNA insertion.
Finally, we shall sequence the isolated cDNA and genomic RAT clones to confirm that they correspond to the junction region and search various data bases for ORFs and map positions on the Arabidopsis genome (three of the RAT genes that we have cloned so far mapped to sequenced regions of the genome, making subsequent genetic mapping unnecessary). Using data bases, we have been able to assign a likely identity or function to many of the RAT genes isolated in our laboratory (Table 1). However, if a RAT gene has not been previously sequenced and mapped, we shall sequence its cDNA and genomic clones and map it using recombinant inbred lines (51,76). We have been successful in this approach with other rat mutants (Nam and Mysore et al., in preparation; Table 1).
6. Complementation of rat mutations by wild-type RAT genes
To prove that the identified cDNA represents a RAT gene, we will introduce the isolated wild-type RAT cDNA into the corresponding rat plant and test its ability to rescue the mutant phenotype, i.e., make it susceptible to the Agrobacterium transformation. Although such complementation analyses would appear to be straight-forward, we foresee two major experimental complications that may have to be resolved.
The first potential complication is, how does one genetically transform a mutant plant that was originally identified as transformation deficient? Although all the rat mutants that we have identified to date cannot easily be transformed by infecting cut roots or stems, they are all highly transformable using flower vacuum infiltration. (This result suggests that vacuum infiltration and wound inoculation may use somewhat different transformation mechanisms or are organ-specific; Mysore et al., submitted). Thus, we shall use flower vacuum infiltration to introduce the wild-type RAT genomic or cDNA clones (under the control of the CaMV 35S or "super-promoter") into the mutant plants. The T-DNA binary vectors containing RAT genes will also carry a gene for hygromycin-resistance. We shall select hygromycin-resistant transformants and test them for tumorigenesis using the assays described above. We expect that, because of different expression levels of the wild-type RAT transgene, not all transformants may fully restore the wild-type phenotype. However, restoration to 10-20% tumorigenesis is significant if the original mutant were transformed at the rate of only 0-5% (this is the situation with all rat mutants identified to date). We shall assure that the complementation does not result from "environmental" or "physiological" effects by testing the co-segregation of the restored phenotype with the complementing T-DNA (i.e., hygromycin resistance).
The second complication may occur with dominant rat mutations. Complementation may then be difficult if not impossible. In fact, many of the rat mutations that we have identified to date are dominant in the diploid plant. (Because the T-DNA did not insert into the coding regions of the genes, we know that these are not dominant negative mutations). Still, we have been able to complement some of these mutations using strong promoters to drive wild-type RAT gene expression, suggesting that overexpression of the wild-type gene can complement even the apparently dominant rat phenotypes. Thus, we think that many of our rat mutants show a dosage effect upon gene expression, and that half the expression level falls below some critical threshold level of expression (haplo-insufficient mutants).
For some RAT genes, the cDNA transgenes may not recreate the particular spatial and/or temporal expression patterns potentially required for the RAT protein function. In such case, we plan to use a native copy of the gene derived from the genomic DNA clone (see above) to rescue the rat phenotype. This genomic fragment is expected to contain the "native regulatory" sequences and recreate the natural expression pattern of this gene. Generally, genomic clones have been used successfully to complement many Arabidopsis mutations (see, for example, 79).
Finally, if these conventional approaches fail, we shall use antisense and/or sense-cosuppression technologies (61,66) to inactivate the wild-type RAT gene using the identified cDNA sequences. Recreation of the rat phenotype in the plants transgenic for the antisense or sense-cosuppressing construction will indicate that the cloned gene indeed encodes the RAT function.
7. Characterization of rat genes
Once we have identified genes involved in the rat phenotype, we shall begin investigation of their roles in the transformation process. We predict that we shall identify genes involved in cell wall biosynthesis and structure (deficient in binding), in DNA repair and recombination (deficient in T-DNA integration), and T-DNA nuclear import. Characterization of each rat gene in detail, however, is beyond the scope of this proposal.
Note that some genes involved in the interaction with Agrobacterium may be difficult to identify using the genetic assay for rat mutants because a deficiency in these gene products may be lethal to the plant. We hope to isolate these "essential" genes using the yeast two-hybrid and one-hybrid systems, RNA differential display, and DNA microarrays.
(Return to top of document)
B. Identification of Arabidopsis proteins that directly interact with Agrobacterium Vir proteins in yeast one- and two-hybrid systems
In host-pathogen interactions, the infecting microorganism generally does not invent novel metabolic pathways; instead, it usurps existing cellular processes and adapts them for use in its life cycle. Thus, Agrobacterium likely employs endogenous plant cell pathways for protein and nucleic acid transport across cellular and nuclear membranes and for integration of T-DNA molecules into the nuclear genome. It may be possible to discover components of these pathways by identifying plant-encoded proteins that directly interact with Agrobacterium proteins. We shall use two experimental approaches to achieve this goal.
1. Use of two-hybrid assay
As mentioned in the Introduction, VirF, VirB2 (and, possibly, VirB3 and VirB9), VirD2, and VirE2 proteins may function within or in direct contact with the host plant cell. These Vir products, therefore, most likely are recognized by and/or interact with plant cellular proteins. We plan to identify putative VirF-, VirB2-, VirB3-, VirB9-, VirD2-, and VirE2-interacting proteins using the yeast two-hybrid assay for protein-protein interaction (28). To this end, we have constructed two-hybrid vectors that express VirB2, VirD2, and VirE2 fused to the Gal4 DNA binding domain. Presently, we are preparing similar constructions for VirF, VirB3, and VirB9 expression. Furthermore, we have also made a two-hybrid cDNA expression library from total seedlings of Arabidopsis (ecotype Columbia) which comprises 5x106 independent cDNA clones with an average size of 1.5 kbp in the plasmid pGAD424 (Clontech). We have already used this library in a two-hybrid assay to isolate an Arabidopsis protein, AtKAPalpha, that mediates nuclear import of VirD2 (14). In addition, we have obtained three other two-hybrid Arabidopsis libraries prepared by the Walker and Theologis laboratories (ARBC stock numbers CD4-10 and CD4-22, respectively) and the Goodman laboratory.
We shall screen the various Arabidopsis two-hybrid libraries with VirF, VirB2, VirB3, VirB9, VirD2, and VirE2 proteins as baits. Each screening process will consist of three steps: (i) growth on a drop-out medium following induction of a selectable gene required for prototrophy; (ii) screening of the resulting colonies for the expression of a reporter enzyme, beta-galactosidase; and (iii) elimination of false-positive clones using non-specific baits (4,5). The identified cDNAs will be sequenced and used as probes to isolate the corresponding full-length clones from standard Arabidopsis cDNA libraries available in our laboratories (4; Tao and Gelvin, in preparation).
We shall test proteins identified in the two-hybrid experiments for the ability to interact with their corresponding ligands using an independent coimmunoprecipitation assay. We have successfully used this approach to characterize the interaction between VirD2 and AtKAPalpha (4). Briefly, we shall clone the identified Arabidopsis cDNA as well as the ORFs of their respective Vir protein ligands into the vector pGEM7Zf+ and use them as templates for in vitro transcription. Messenger RNAs transcribed with T7 DNA polymerase and capped with 7-methyl-guanosine will be translated in vitro using a wheat germ translation system supplemented with 35S-methionine (4,58). Translation mixtures will be combined and incubated to allow interaction. After incubation, anti-Vir protein antibodies, already available in our laboratories, will be added, followed by addition of a protein A bead slurry and centrifugation. After washing, proteins will be analyzed by SDS PAGE (49) and autoradiography. If the identified plant cDNA represents a specific interactor, its translation product will coprecipitate only with the corresponding Vir protein.
The proposed two hybrid experiments are expected to identify genes involved in four different biological processes:
a. VirF-interacting proteins will likely reveal one molecular mechanism by which Agrobacterium host range is determined.
b. VirB-interacting proteins will probably represent host factors involved in docking of the bacterial pilus at the cell surface, penetration of the cell membrane by the pilus and/or transport of the T-complex into the host cell through the VirB channel.
c. VirD2- and VirE2-interacting factors will include those involved in nuclear import of T-complexes. We have already identified two Arabidopsis proteins (AtKAP alpha and PP2C) that specifically bind the VirD2 NLS and may mediate and/or regulate its nuclear uptake (4; Tao and Gelvin, in preparation). However, no host proteins that recognize the NLS signals of VirE2 have yet been identified. Isolation of such a VirE2-interacting protein is an especially interesting objective because VirE2 nuclear import is plant-specific (35; see also below).
d. Both VirD2 and VirE2 are multifunctional proteins that, in addition to nuclear import, most likely participate in integration of the imported T-DNA (7,63,83,91). Thus, VirD2- and VirE2-interacting proteins likely will include host factors involved in DNA repair and recombination.
2. Use of one-hybrid assay
This approach takes advantage of our observations that the nopaline-type VirE2 NLS is active in plants but not in animal or yeast cells (see Preliminary Results and 35). Potentially, VirE2 nuclear import in yeast may be complemented by expression of plant cDNA clones that encode a putative VirE2 NLS receptor. To identify such cDNAs, we have developed a one-hybrid system that allows specific selection of yeast cells in which VirE2 is transported into the cell nucleus. In this technique, a triple-fusion protein composed of LexA, the Gal4 activation domain, and VirE2 is expressed in the yeast strain L40. If imported into the nucleus, LexA targets the fusion protein to LexA operator sites upstream of the lacZ and HIS3 reporters and Gal4 domain activates the expression of the reporter genes, resulting in yeast growth on a histidine-deficient medium as well as in beta-galactosidase activity.
The key component of the vector pLGE2 is a modified LexA gene (mLexA) which was produced in one of our laboratories (VC). The success of this nuclear import assay hinges on the inability of a LexA::Gal4 fusion protein to enter the nucleus in the absence of a NLS. Thus, neither LexA nor Gal4 should contain NLS sequences. Indeed, the Gal4 activation domain is known to lack a NLS whereas LexA, a bacterial protein, is generally thought not to have evolved such a signal. However, we found that wild-type LexA carries a previously unidentified NLS sequence, rendering the above described experimental design impossible. To circumvent this difficulty, we identified the LexA NLS and inactivated it by specific substitutions of two amino acid residues, R157G and K159E.
We constructed two variations of the triple fusion protein, with and without the SV40 NLS sequence. In the presence of SV40 NLS, the fusion protein entered the cell nucleus and activated lacZ gene expression, resulting in beta-galactosidase activity. In the absence of the SV40 NLS, the fusion construction was unable to reach the cell nucleus and, thus, did not activate the reporter gene (data not shown). Thus, the mLexA:Gal4:VirE2 fusion activated expression of both reporter and selection genes and this activation depended on a functional NLS signal. In addition, our previous studies have demonstrated that fusing other proteins to VirE2 did not affect its NLS function (18,20), suggesting that the plant-specific VirE2 NLSs will remain active in the context of the mLexA:Gal4:VirE2 fusion.
Here we shall use this one-hybrid system to screen an Arabidopsis cDNA library in the constitutive URA3+ yeast expression vector pFL61 (62; kindly provided by F. Lacroute, CNRC) for clones that support VirE2 nuclear import in yeast cells. Following transformation, we shall plate the cells on a uracil, histidine, and tryptophan triple drop-out medium. Transformants prototropic for all three nutrients and expressing beta-galactosidase activity will likely represent cells in which mLexA:Gal4:VirE2 was redirected to the nucleus to activate HIS3 and lacZ expression. cDNA clones from these surviving cells will be isolated and sequenced.
(Return to top of document)
C. Identification of Arabidopsis genes induced or repressed during the initial stages of Agrobacterium transformation
To date, the potential effects of Agrobacterium-plant cell interactions on host gene expression have not been adequately addressed. We are aware of only one report (10) that investigated differences in gene activity following Agrobacterium transformation. However, this study described differences in mRNA between potato plants and mature potato crown gall tumors. Here, we propose to identify Arabidopsis genes induced or repressed during the early stages of Agrobacterium infection, i.e. from the initial infection up to the integration event. To this end, mRNA will be prepared from uninfected and Agrobacterium-infected Arabidopsis cell cultures. We have previously shown that 20% to 60% of the plant cells in a suspension culture can be transiently transformed by Agrobacterium that have been pre-induced with acetosyringone (63; N. Darbinian, L.-Y. Lee, and S. Gelvin, unpublished observations). The infected cells will be first sampled immediately after infection and then at various time periods (e.g. every 6 hours) up to 24 hours when T-DNA integration is thought to occur (63,104). We shall subsequently use three experimental techniques to analyze differential gene expression in these mRNA preparations.
1. Use of subtraction techniques
We shall construct cDNA subtraction libraries using mRNA from Arabidopsis cell cultures infected with a "disarmed" Agrobacterium strain (GV3101) containing the T-DNA binary vector pBISN1 (67), and a similar bacterial strain that contains pBISN1 but lacks vir genes. This latter strain cannot transfer T-DNA, but will act as a "mock-infection" control for non-specific Agrobacterium-induced "stress effects" upon the cell culture. We shall prepare the "infected" cDNA with EcoRI cohesive ends while we shall make the "mock-infected" cDNA blunt-ended. We shall further digest the blunt-ended cDNA preparation with the restriction endonucleases RsaI and AluI, resulting in a large number of small blunt-ended DNA fragments. We shall next mix and anneal the "infected" cDNA preparation with a 50-fold (w/w) excess of the "mock-infected" digested cDNA. We shall use the annealed cDNA inserts to construct a library in an EcoRI-digested vector. The only cDNA clones likely to be inserted into the vector are those sequences that are double-stranded and have EcoRI cohesive ends. These clones are derived from the "infected" cDNA preparation and represent sequences for which no complementary fragments were present in the "mock-infected" cDNA, resulting in a subtraction library enriched for genes induced by Agrobacterium infection. To enrich for genes suppressed by the infection, the procedure will be reversed, using "mock-infected" cDNA with EcoRI ends and "infected" cDNA with blunt ends.
To identify rare differentially-expressed messages, we plan to utilize a PCR-based procedure routinely used in our laboratory. In this approach, we shall amplify the subtracted cDNAs by two rounds of PCR using a commercially-available PCR-Select� kit (Clontech); this procedure is fast (takes only 3-4 days) and requires small amounts of starting material (0.5-2.0 mu g of polyA+ RNA).
2. Use of RNA differential display
Alternatively, we shall perform RT-PCR to display subsets of mRNAs as short DNA bands (50). Such mRNA fingerprinting will allow us to observe alterations in Arabidopsis gene expression caused by different environmental factors, such as Agrobacterium infection. The resulting DNA fragments of interest can easily be re-amplified, cloned, and sequenced. In addition, these clones can be used as hybridization probes to identify genes from genomic or cDNA libraries.
3. Use of DNA microarrays
We shall produce microarrays of Arabidopsis EST cDNA clones using the microarrayer at Purdue University (see Facilities statement below). Alternatively, we shall purchase Arabidopsis cDNA microarray chips from the NSF genomics center at Michigan State University (see attached letter). We shall screen these DNA chips (84) with probes prepared from mRNA from "mock-infected" and Agrobacterium-infected Arabidopsis cell cultures using the robotic reader in the Biotechnology Facility at Stony Brook, or the microarray reader at the NSF genomics center at Purdue.
Of critical importance to this analysis will be the choice of cDNA clones to use for microarray analysis. Obviously, we shall utilize as many different cDNA clones as possible, but we may be limited by the number of cDNAs that we can physically handle and the availability of a "complete" cDNA library. We hope to analyze at least 10,000 different genes. We shall necessarily include genes that we have identified by the rat mutant screening and the yeast one- and two-hybrid analyses described above. We shall also microarray genes (and gene families) that are "related" to those genes identified above. For example, we have identified a specific histone H2A gene as essential for Agrobacterium-mediated transformation. We shall therefore include for microarray analysis other histone and chromatin-related genes.
In this part of the proposed research, we expect to identify Arabidopsis genes that are either induced or repressed early during the Agrobacterium transformation process. To focus further on the specificity of this differential gene expression, we shall include the following experimental modifications and controls:
a. To avoid detecting expression of Arabidopsis phytohormone response genes contained within a wild-type T-DNA, we shall use a disarmed (non-tumorigenic) Agrobacterium strain.
b. To control for non-specific mutagenic effects of the integrated T-DNA on host gene expression, we shall use RNA derived from hundreds of thousands of Arabidopsis cultured cells. Our focus on the early stages of the infection should help avoid long term effects of T-DNA integration.
c. To avoid cross-hybridization with different members of the same multigene family, we shall use PCR-generated microarray sequences that are gene-specific rather than whole cDNAs.
d. To control for non-specific general stress and defense responses, we shall incubate the "uninfected" control cell cultures with an Agrobacterium strain that lacks vir genes. This strain thus cannot transfer T-DNA to the plant cells. Any induction or repression of Arabidopsis genes resulting from incubation with this "non-infectious" Agrobacterium strain will therefore not represent a plant response to T-DNA transfer, nuclear targeting, or integration.
(Return to top of document)
D. Biological characterization of Arabidopsis genes involved in transformation by Agrobacterium
Elucidation of the biological activity of Arabidopsis genes and proteins identified in this research is critical for understanding their role in the process of Agrobacterium-plant cell interactions. Here, we shall address this question using four experimental approaches.
1. Use of reverse genetics
We shall utilize a reverse genetics approach to investigate Arabidopsis genes that may be involved in Agrobacterium-mediated transformation. Targets for this investigation will include three categories of genes: (i) Additional members of multigene families for which we have already identified a rat gene by direct screening of T-DNA insertion collections; (ii) Genes identified by the yeast one- and two-hybrid systems that are likely to interact with Vir proteins that are present on the bacterial surface or that are transported into the plant together with the T-complex; (iii) Genes identified by subtraction, differential display, and microarray analyses that are up- or down-regulated specifically upon Agrobacterium infection of plant cells.
a. Identification of rat Mutants in Multigene Families. Krysan et al. (45) recently described a PCR protocol to identify mutants in known gene families found in the Feldmann and DuPont T-DNA insertional mutagenesis collections. Briefly, DNA is amplified from pools of 100 T-DNA mutagenized plants using one primer directed to a T-DNA border and one primer directed to a region of known sequence within the gene of interest. DNA from amplified bands is excised and used as a hybridization probe to demonstrate a RFLP between the wild-type and mutant plants. The pools of 100 plants are divided into 5 pools of 20 plants and the PCR reaction performed again. Once the pool of 20 plants containing the T-DNA insertion has been identified, individual plants are tested.
We propose to perform similar analyses to isolate Arabidopsis mutants in members of already identified RAT multigene families. To this end, we shall use PCR primers directed against the left and right T-DNA borders (because of T-DNA rearrangements and the generation of head-to-head dimers during T-DNA insertion, it is unpredictable whether a disrupted gene will contain a T-DNA containing both left and right borders, two left borders or two right borders as junctions) and a primer directed to the gene of interest. For example, we have identified a T-DNA insertion mutant (rat4) containing a disrupted CSL gene (Table 1). To determine whether other CSL or related CELA genes are involved in Agrobacterium-mediated transformation, we shall design a PCR primer to one of the conserved UDP-glucose binding domains within these genes (74) and identify csl and celA mutants in the various T-DNA insertion collections. We shall subsequently test these mutants for the rat phenotype. Another set of target genes will be the histone H2A gene family. We have already identified a histone H2A variant (rat5) involved in the T-DNA integration process (Mysore et al., in preparation). As we identify other rat mutants in multigene families, we shall target other members of these families for mutagenesis using this reverse genetics approach.
b. Identification of Mutants in genes Isolated Using Yeast One- and Two-Hybrid Systems and Subtraction, Differential Display, and Microarray Analyses. We shall identify additional targets for "reverse genetics" from the yeast one- and two-hybrid experiments. For example, using the two-hybrid system, we have identified an Arabidopsis AtKAP alpha gene encoding one of the proteins, karyopherin alpha, that participate in T-DNA nuclear import (4). AtKAPalpha (Accession Number U69533) belongs to a multigene family which includes aIMPalpha (Accession Number AF077528) and AtIMPalpha1-4 (Accession Numbers Y14615, Y14616, Y15224, and Y15225, respectively). Potentially, inactivation of AtKAPalpha in Arabidopsis mutants will not completely block nuclear import but may lead to the development of a specific phenotype. Similarly, we have identified a tomato Type 2C protein phosphatase (PP2C) that is likely involved in regulating nuclear import of T-DNA (Tao et al., in preparation). We shall identify AtKAPalpha and Arabidopsis PP2C gene "knockouts" and determine their effect on nuclear import of VirD2 using transient expression of GUS-VirD2 constructions (18,38) as well as Agrobacterium tumorigenicity assays (20). Or, Arabidopsis proteins that interact with the VirB2 pilin protein will likely be involved in bacterial attachment to the plant cell surface. A lower level of expression of this plant protein may result in decreased bacterial attachment (64,65).
Finally, we shall isolate Arabidopsis mutants in up- or down-regulated genes that we identified using the subtraction hybridization, RNA differential display, and microarray approaches. These genes were identified only as being associated with the early steps of the Agrobacterium transformation process. The reverse genetics approach will indicate whether they are essential for it.
We would like to note that we may not be able to identify "knockouts" of genes that are essential for plant growth and development. However, we have already determined that many genes involved in Agrobacterium infection belong to multigene families. For example, AtKAPalpha (Accession Number U69533) belongs to a multigene family that includes aIMPalpha (Accession Number AF077528) and AtIMPalpha 1-4 (Accession Numbers Y14615, Y14616, Y15224, and Y15225, respectively). Potentially, individual members of such multigene families are not critical for plant survival but are required for Agrobacterium-mediated transformation. Furthermore, most of the T-DNA insertion rat mutants isolated to date are not "knockouts" per se because the T-DNA had inserted into the 5' or 3' untranslated regions of the RAT genes. These may therefore represent "knockdown" mutants that express lower levels of the gene products, allowing the plant to survive but not permitting Agrobacterium-mediated transformation.
Technically, the reverse genetics experiments will be conducted in two ways. We have obtained the Feldmann and DuPont collections and have set up a "reverse genetics" facility at Purdue based upon the approach described above. We shall expand this facility to screen the Bressan collection of T-DNA insertion mutants that is currently being generated at Purdue. Also, we shall take advantage of the screening service that is currently being developed by Dr. Michael Sussman (Univ. of Wisconsin) using the Amasino/Sussman collection of T-DNA insertion mutants. As mentioned above, the combined collections from Feldmann, DuPont, Bressan, and Amasino/Sussman saturate the entire Arabidopsis genome. Thus, the reverse genetics approach proposed here will likely discover mutations in most genes identified in this study.
2. Use of nuclear import assays
VirD2- and VirE2-interactors may be involved in nuclear transport of these Agrobacterium proteins and, by implication, the T-complex itself. We shall test this hypothesis four ways:
a. Quantification of Nuclear Import in Plant Protoplasts. We have devised an assay to quantify the extent of nuclear translocation of a GUS-VirD2 NLS fusion protein in tobacco BY-2 protoplasts (Tao and Gelvin, in preparation). This assay can be easily adapted for use in Arabidopsis protoplasts. Briefly, a gene encoding a nuclear import substrate, i.e., GUS-VirD2 or GUS-VirE2 fusion proteins, is electroporated into protoplasts and the cells are stained with X-gluc 24 hours later. The percentage of cells showing exclusive nuclear localization of GUS activity (usually 80%) is determined. Next, the gene encoding the NLS-interacting protein to be tested is co-electroporated into the cells with the nuclear import substrate gene and the extent of GUS activity localized exclusively in the nucleus is determined. Proteins (such as PP2C described above) having a negative effect upon nuclear localization will lower the percentage of co-transfected cells showing exclusive nuclear localization of GUS activity. Proteins that have a positive effect upon nuclear localization will raise this percentage. Alternatively, we can express an anti-sense construction to genes encoding proteins that positively effect nuclear localization. Such expression should lower the percentage of co-transfected cells showing exclusive nuclear localization of GUS activity.
b. Binding to VirE2 NLS. No plant proteins that recognize the VirE2 NLS have been identified to date. To examine whether VirE2-interacting protein(s) specifically recognize its NLS signal, we shall perform in vitro translation and coimmunoprecipitation experiments as described above using VirE2, the VirE2 interactor, and anti-VirE2 antibodies. If the VirE2-interacting protein indeed represents a specific NLS receptor, its translation product will coprecipitate with the wild-type VirE2 protein but not with VirE2 derivatives lacking the functional NLS sequences (18, 20).
In addition, we shall use coimmunoprecipitation to examine whether the VirE2 NLS receptor also recognizes the NLSs of VirD2. Although AtKAPa was unable to bind VirE2 (4), it is possible that the VirE2 NLS receptor has a more broad affinity, interacting with both plant-specific (VirE2) and general types (VirD2) of NLS signals. If both VirE2 and VirD2 interact with the VirE2 NLS receptor, the affinity of this interaction will be examined by adding increasing amounts of salt to the immunoprecipitation reactions. The salt concentration required to dissociate protein-protein or protein-DNA complexes is commonly used to estimate binding affinity; usually, dissociation by 0.05-0.2 M of NaCl is considered a low affinity interaction whereas dissociation by salt concentrations of 1.0-1.5 M represents a strong binding (13, 17).
c. VirE2 nuclear import in Animal and Yeast Cells. We hypothesize that the VirE2 NLSs do not function in animal systems because these cells lack the NLS receptors that recognize the VirE2 nuclear targeting sequences. Thus, we expect that introduction of such receptors into Drosophila embryos or Xenopus oocytes will promote nuclear uptake of VirE2 and VirE2-ssDNA complexes in these cells. To test this notion, the plant NLS receptor found to interact specifically with VirE2 will be mixed with fluorescently-labeled VirE2 protein (or VirE2-ssDNA complexes) and incubated for 15-30 minutes to allow formation of VirE2-NLS receptor complexes. These complexes will then be microinjected into Drosophila embryos or Xenopus oocytes as described (35). Nuclear uptake will confirm that we have identified a plant-specific NLS receptor responsible for VirE2 nuclear import.
By analogy with non-plant systems (3,41,47), VirE2 nuclear import may involve general posttranslational modifications (e.g. phosphorylation) of the NLS receptor. These modifications are more likely to occur during protein expression in living cells than immediately after microinjection. Thus, in addition to microinjection, VirE2 protein and its NLS receptor will be coexpressed in yeast cells (4). Following expression, VirE2 nuclear import will be assayed by immunofluorescence using anti-VirE2 antibodies. This approach will expand our original one-hybrid experiments. Here, we will use wild-type VirE2 rather than the mLexA:Gal4:VirE2 fusion as nuclear import substrate and monitor its nuclear accumulation directly by immunofluorescence microscopy.
d. Functional Study of the Putative VirE2 NLS Receptor In Vitro. Protein microinjection or expression in living cells as an assay for nuclear transport has some limitations. For example, other cytosolic components may interfere with the process of nuclear import, complicating interpretation of the experimental data. The use of an in vitro nuclear import assay circumvents these difficulties (reviewed in 34). Presently, however, no plant experimental system exists in which the role of isolated components of protein nuclear import machinery can be tested in vitro; nuclear import of NLS-containing proteins in permeabilized protoplasts of different plant species has been shown to occur without the addition of plant cytosol (8,36,59). Thus, we plan to use permeabilized yeast cells (86) to examine the function of the purified plant karyopherins in vitro. We have already utilized this system to study AtKAPa (4). Here, we shall perform the assay using the isolated components sufficient to detect nuclear import (102). Specifically, we shall include in the reaction mixture import substrates (i.e. fluorescently labeled VirD2, VirE2 or VirE2-ssDNA complexes), permeabilized yeast cells depleted of their cytosol (86), and the recombinant karyopherin beta and Ran GTPase proteins (both kindly provided by Colin Dingwall, Stony Brook). We shall add the VirE2 NLS receptor to the reaction and monitor and record nuclear uptake with a confocal microscope. We shall further assess the specificity of interaction with the NLS signals using previously generated VirD2 and VirE2 derivatives with mutated NLS sequences (4,18,20,35). We shall purify all tested proteins after overexpression of their cDNAs in E. coli (4,36).
3. Use of in vitro integration assays
T-DNA integration is thought to occur by a multistep process including: (i) recognition of partially double-stranded chromosomal breaks in the host DNA by VirD2 covalently attached to the 5' end of the T-DNA, (ii) search for microhomologies within the host genomic DNA by the 3' end of the T-DNA; this reaction may be assisted by VirE2; (iii) trimming of non-homologous DNA ends by cellular exo- and endonucleases, and (iv) joining of the DNA ends by VirD2 with the help of cellular ligases and other protein functions involved in illegitimate recombination (31,57,83,97). In addition, general properties of dynamic chromatin, such as accessible chromatin structure, availability of a nicks, and transient single-stranded regions in the chromosomal DNA, are expected to be required for efficient T-DNA integration.
Here, we shall subject extracts from Arabidopsis rat mutants tentatively characterized as integration defective in the described above genetic and biochemical analyses to two in vitro tests to determine where the multistep process of integration may be impaired.
a. In vitro Integration. This approach, recently developed in one of our laboratories (BH), will test cell free nuclear extracts from young Arabidopsis seedlings prepared as described for extraction of pea shoot apices or tobacco BY-2 cells (21,90). We shall produce the partially double-stranded integration target by annealing the 5'-radiolabeled 13-mer oligonucleotide 5' TAGCCAAACGTAC 3' to the unlabeled oligonucleotide 5' CTGACTGGGTACGTTTGGC 3' at a molar ratio of 1:1, resulting in the following DNA fragment (* indicates the 5'-end radiolabel):
5'* TAGCCAAACGTAC 3'
3' CGGTTTGCATGGGTCAGTC 5'
We shall obtain the single-stranded integration substrate, i.e., a VirD2-T-DNA complex, by reacting the oligonucleotide 5' GCTCAAATTACAACGGTATATATCCTG^CCAGTCAG 3' (the border sequence is underlined and the VirD2 cleavage site within it is marked with ^) with VirD2 purified following expression in E. coli (72). In this reaction, VirD2 will cleave the border and covalently attach to the 5' end of the nick, resulting in the following complex: VirD2-5' CCAGTCAG 3'. For the integration test, we shall mix the target (4 pM) and the substrate (8 pM) with the plant extract (5-20 mu g protein), 20 mM Tris-HCl pH 8.8, 50 mM NaCl, 5 mM MgCl2 5 mM ATP and incubate the mixture for 15 minutes at room temperature. Integration of the VirD2-oligonucleotide complex will generate the following 21-mer radiolabeled integration product: 5'* TAGCCAAACGTAC"CCAGTCAG 3' (the ligation junction between the two integrated oligonucleotides is indicated with "). We shall stop the reaction with formamide (30% final concentration) and separate the products on 20% polyacrylamide/8 M urea gels. We shall calculate the efficiency of integration as the ratio of the radioactivity in the labeled integration product (21-mer) to the total radioactivity in the reaction (13-mer and 21-mer). This assay will determine whether the tested plant is impaired in the events specifying integration of the 5' end of the T-DNA.
b. DNA End-Joining Test. Second, we shall analyze the Arabidopsis extracts in a test that examines the efficiency of ligation of the T-DNA 3' end to non-specific target DNAs. As a model target DNA, we shall utilize a partially double-stranded oligonucleotide containing a free 5' end and limited homology to the T-DNA 3' end (that includes 22 of the 25 bases of the border sequence). This assay will determine whether the tested plant is impaired in the cellular activities specifying integration of the 3' end of the T-DNA. By analogy to yeast and animal systems, protein functions potentially involved in 3' end-joining reaction are ligases and their interacting proteins (98) and components of the DNA-PK (6).
Several modifications of the proposed assays will be included in later stages of the work:
a. We shall test proteins interacting with VirD2 and VirE2 isolated in the two-hybrid assays (except for NLS binding proteins) using the described above assays.
b. We shall use antibodies to these VirD2 and VirE2 interactors to block the reactions, confirming the role of these proteins in the integration process.
c. We shall increase the length of target DNAs as well as of VirD2-oligonucleotide complexes to allow better analysis of the potential requirements for sequence homology during integration.
d. We shall add purified VirE2 to the longer VirD2-oligonucleotide complexes to mimic the structure of the natural T-complex. This will enable us to analyze the possible influence of VirE2 on integration; for example, the tightly packaged telephone cord-like structure into which the ssDNA is shaped by VirE2 (16) may help to preserve the integrity of the integrating DNA (83).
e. We shall repeat these experiments using extracts from maize plants.
Overall, the experiments proposed in this section will not only help in the analysis of rat mutants and individual Arabidopsis proteins, but will lead us to a deeper understanding and possible manipulation of the process of T-DNA integration.
4. Intracellular localization studies
Identification and/or characterization of Arabidopsis genes and their products by functional assays, e.g. nuclear import and recombination and integration, will likely indicate their possible role in Agrobacterium infection. Most likely, these factors will represent VirD2- and VirE2-interacting proteins. In contrast, genes identified in the two-hybrid experiments with VirF and VirB proteins as baits as well as differentially expressed genes may be more recalcitrant to functional characterization.
One way to address this problem is to determine the exact intracellular localization of the examined protein in healthy as well as in Agrobacterium-infected Arabidopsis. This information may provide the first insight into the biological function of the studied protein. For example, proteins localized to the host cell membrane may be involved in recognition of Agrobacterium, determination of its host range, and/or interaction with the VirB pilus. Cytoplasmic proteins may be involved in transport of the invading T-complex from the VirB channel to the cell nucleus. Nuclear proteins, on the other hand, will likely participate in the integration event. Finally, the use of Agrobacterium-inoculated plants or cell suspensions may even detect potential colocalization of some of the bacterial proteins and/or structures with the identified cellular factors. Although this approach may not definitively determine the biological function of the tested proteins, it will point us in the right direction, allowing us to design future experiments to address directly the role of the identified proteins in the Agrobacterium infection process.
Technically, we shall use immunoconfocal and immunoelectron microscopy to determine intracellular locations of the tested proteins. For immunoconfocal microscopy, tissue sections will be stained with primary antibodies followed by an inodicarbocyanine (Cy5)-conjugated secondary antibody. Cy5 is excited near 650 nm and fluoresces near 670 nm, circumventing the autofluorescence associated with plant tissues. Although Cy5 cannot be seen well by eye and therefore is unsuitable for conventional fluorescence microscopy, it is excited with Krypton/Argon and Helium/Neon lasers used in confocal microscopy. We have used this approach to immunolocalize turnip vein clearing virus (TVCV) particles in TVCV-infected tobacco plants (32). For immunoelectron microscopy, we shall use a protocol recently developed in our lab (32). Most immunoelectron microscopy techniques significantly compromise cellular membranous structures as well as overall contrast and resolution of the image. Our procedure, based on OsO4 post fixation and embedding in Durcupan ACM resin, avoids these flaws but retains full antigenicity of the sample (32).
(Return to top of document)
E. Improvement of agronomically important crops using Arabidopsis genes essential for Agrobacterium infection
Identification and isolation of Arabidopsis genes encoding proteins that participate in Agrobacterium-plant cell interactions will set the stage for one of the ultimate goals of all Agrobacterium research: The improvement of agronomically and horticulturally important crops by making them susceptible to efficient genetic manipulation using Agrobacterium as a gene transfer vector. Conversely, there is a pressing need to genetically modify certain commercial plant species that suffer losses resulting from Agrobacterium infection. Important plants to target for crown gall resistance include grape, apple, stone fruits, walnut, rose, and Chrysanthemum (see, for example, 9). Finally, disruption of the predominant mechanism of T-DNA integration by illegitimate recombination may allow us to increase the frequency and/or recovery of genes introduced by homologous recombination. (The use of integration-deficient rat mutants to effect homologous recombination is the subject of a grant to SBG funded by the Consortium for Plant Biotechnology Research. Thus, we shall not include these experiments in this proposal).
The first three years of work described in this proposal will focus upon the identification of Arabidopsis genes involved in the Agrobacterium transformation process. However, in the fourth and fifth years we shall make a major effort to apply the knowledge that we have gained for the genetic improvement of important crop species. Although there are numerous agronomically important plant species that are currently recalcitrant to Agrobacterium-mediated genetic transformation, we shall focus upon maize. We have chosen this species because of its immense agronomic importance, the availability of technology for plant regeneration, and the knowledge that at least some varieties can be transformed by Agrobacterium. Similarly, we shall focus on grape as a species in which to develop crown gall resistance. We have chosen this species because of the importance of crown gall disease in viticulture, and the availability of an Agrobacterium-based transformation and regeneration system.
1. Manipulation of maize to render it more susceptible to transformation by Agrobacterium
Although Agrobacterium-mediated transformation of maize has been achieved (39), high-efficiency genetic modification of this species is currently restricted to particular genotypes (especially A188 and crosses that contain A188 as a parent). Many agronomically elite varieties remain recalcitrant to Agrobacterium-mediated transformation. Furthermore, the increasing evidence suggests that, at least using these maize varieties and transformation protocols, T-DNA integration may be an important block point in the transformation process. Indeed, transient transformation of maize is relatively efficient and has been obtained by many groups (see, e.g., 67,78,89) whereas the stable genetic transformation of maize using Agrobacterium has been more problematic. As a starting point to improve the stable transformation of maize, we shall employ two different approaches:
a. Expression of Arabidopsis "Integration-Related" Genes in Maize. We shall introduce Arabidopsis genes that we have identified as important for T-DNA integration into the maize genome. We first shall modify genetic regulatory elements associated with these genes to optimize expression in monocots. These modifications will include using promoters that function efficiently in maize (such as the maize ubiquitin or actin promoters, or our "super-promoter" that we have recently determined works well in maize (Kononov et al., in preparation), and a maize adh1 intron that increases gene expression levels in monocots (11). Because the elite varieties of maize that we shall transform are currently recalcitrant to Agrobacterium-mediated transformation, we shall use particle bombardment. Previously, one of us (SBG) has contracted with the maize genetic transformation facility at Iowa State University. This facility generated for him several hundred transgenic maize plants using particle bombardment. We shall again utilize this facility to generate maize plants transgenic for Arabidopsis "integration-related" genes modified for expression in monocots.
We shall determine the expression levels of the modified Arabidopsis transgenes, then conduct Agrobacterium-mediated transformation experiments on lines that express these genes well. We shall compare (using gusA or gfp genes) transient and stable transformation frequencies between the transgenic maize and the non-transgenic parent lines. In some instances, we shall introduce several "integration" genes into the maize genome, either by incorporating multiple genes during bombardment or by crossing maize plants containing different transgenes. We shall correlate stable transformation with the number, type, and expression levels of the "integration" transgenes.
b. Overexpression of Maize Homologues of Arabidopsis "Integration-Related" Genes in Maize. It is possible that low levels of transgene integration into the maize genome result from low levels of expression of endogenous maize proteins important for integration to occur. We shall therefore first identify maize homologues (from the better-transformable cultivar A188) of Arabidopsis genes required for T-DNA integration into the Arabidopsis genome. We shall next examine the expression levels of these genes, especially in cells and tissues (such as scutellar cells; 39) that are readily transformed by Agrobacterium in the maize cultivar A188. We shall clone these genes from maize, then put them under the genetic control of promoters that express strongly in monocots. Finally, we shall introduce these modified putative maize "integration-related" genes into elite varieties of corn using particle bombardment. We shall subsequently determine and correlate the expression levels of these genes in scutellar tissue with the Agrobacterium-mediated stable transformation frequency of these tissues.
2. Manipulation of the grape genome to prevent crown gall disease
Crown gall is an important disease with major economic consequences for the viticulture industry. There is currently no adequate prevention of Agrobacterium vitis infection, nor cure for infected plants and vineyards (9). Therefore, grape remains a major target species for developing crown gall resistance. We shall utilize the information gained in our Arabidopsis studies to develop crown gall-resistant grape. There are several possible steps in the Agrobacterium transformation process that could possibly be blocked to prevent crown gall disease. These include bacterial attachment, T-DNA nuclear targeting, and T-DNA integration. Because inhibition of the nuclear targeting and integration steps may present detrimental side-effects to plant growth and development, we shall first target bacterial attachment. Also, we believe that the earlier the infection process can be blocked, the more likely it is to be effective.
We have already determined that several key cell wall enzymes and structural proteins are important for Agrobacterium attachment to the plant cell surface. Among the structural proteins essential for attachment are an arabinogalactan protein (AGP) that is disrupted in the Arabidopsis rat1 mutant. However, homozygous rat1 mutant plants appear normal in growth and development (Nam et al., in preparation). We shall therefore identify grape homologues of this arabinogalactan protein and target them using anti-sense or co-suppression methodologies. Dr. Tom Burr's laboratory has recently developed efficient grape rootstock transformation technology (Tom Burr, personal communication). We shall work with Dr. Burr's group to develop grape plants with lowered levels of this specific AGP. We shall quantify AGP expression and accumulation levels and correlate this with bacterial attachment and tumorigenesis-susceptibility. As we identify other Arabidopsis genes involved in bacterial attachment, we shall use their homologues as targets for gene disruption in grape.
(Return to top of document)
VI. TIMETABLE
Year 1. Begin genetic assays for rat mutants, identification of differentially expressed genes in Agrobacterium-infected Arabidopsis, and reverse genetics experiments using AtKAPa, histone H2A, AGP, CELA, and CSL genes. Complete screening of Arabidopsis cDNA libraries using the one- and two-hybrid approaches. Start in vitro integration work.
Year 2. Complete identification of differentially expressed genes and isolation of rat mutants. Begin biological characterization of the identified genes and proteins and cloning of RAT genes. Continue integration and reverse genetics work with newly identified Arabidopsis genes.
Year 3. Complete reverse genetics experiments, integration work, and RAT gene cloning. Continue biological assays. At this stage, we expect to obtain enough Arabidopsis genes involved in Agrobacterium infection to allow us to begin using these data for crop improvement.
Year 4. Complete biological assays. Begin experiments to improve Agrobacterium-mediated transformation of maize and to generate crown gall-resistant grape.
Year 5. Complete improvement of Agrobacterium-mediated transformation of maize and the generation of crown gall-resistant grape.
VII. INFORMATICS
This project does not require the development of new informatics systems, nor does it require the use of sophisticated informatics programs. We shall deposit novel DNA sequences in the relevant data bases (e.g., GeneBank), and information describing the rat mutants in the Arabidopsis Mutant data base (Oklahoma State University). In addition, we shall establish a web site describing our research activities and results, and continuously update this information. Ms. Terry Combs (Purdue University) is an expert on establishing such sites, and we shall use her expertise in this endeavor. Specifically, this site will describe details of the genetic and molecular analyses of rat mutants, a list and molecular description of Arabidopsis proteins that interact with various Agrobacterium Vir proteins, and a list and description of Arabidopsis genes that are up- or down-regulated during the initial stages of Agrobacterium infection. We shall also make available a list of mutants that we have obtained using reverse genetics, the primers used in these searches, and a description of the nature of the gene disruption in these mutants. We shall freely distribute all information, bacterial strains, and Arabidopsis mutants obtained during the course of this project. At this point, we do not foresee patenting any of the material developed or information obtained.
(Return to top of document)
VIII. ROLES OF PARTICIPANTS
The proposed research is so extensive and comprehensive that it would present a formidable and virtually unattainable objective for any single laboratory. Only a close collaboration between groups working on the forefront of physiological, genetic, cellular, and molecular aspects of Agrobacterium-plant cell interactions will make this research possible. Stanton Gelvin, the PI, is a plant molecular biologist with special expertise in the genetics of Agrobacterium infection and the early events of the genetic transformation process. Vitaly Citovsky, the U.S. co-PI, is a biochemist and cell and molecular biologist specializing in plant-pathogen interactions and Agrobacterium T-DNA nuclear import. Barbara Hohn, the European co-PI, is a molecular and cell biologist with an unique expertise in the central, yet still elusive, process of T-DNA integration. All three laboratories are well funded, staffed with highly trained postdoctoral researchers, and equipped with all the equipment, including confocal and electron microscopes and DNA microarray robotics, required for the proposed research. Thus, the proposed project has a substantial and unique combination of expertise and infrastructure necessary for its successful completion. The recruitment of students and postdoctoral fellows to perform this research is described in the Appendix.
Table 2. The role of each investigator in the proposed activities
| Stanton Gelvin |
Vitaly Citovsky |
Barbara Hohn |
1. Identification of rat mutants
2. Reverse genetics, microarray
analyses, subtraction analyses
3. In vivo nuclear import assays
4. VirB2 2-hybrid analysis
5. Participate in microarray analysis
6. Gene and phenotype characterization |
1. Identification of rat mutants
2. 1- & 2-hybrid analyses of VirD2 & VirE2
3. In vitro nuclear import assays
4. Differential display and subtraction
analysis
5. Participate in microarray analysis
6. Gene and phenotype characterization |
1. In vitro T-DNA analysis
of rat mutants
2. Gene and phenotype
characterization |
IX. SERVICE COMPONENT
This proposal does not contain a service component.
(Return to top of document)
XI. REFERENCES
1. Arioli, T., L. Peng, A.S. Betzner, J. Burn, W. Wittke, W. Herth, C. Camilleri, H. Hofte, J. Plazinski, R. Birch, A. Cork, J. Glover, J. Redmond, and R.E. Williamson. 1998. Molecular analysis of cellulose biosynthesis in Arabidopsis. Science 279: 672-673.
2. Azpiroz-Leehan, R. and K.A. Feldmann. 1997. T-DNA insertion mutagenesis in Arabidopsis: Going back and forth. TIG 13: 152-156.
3. Azuma, Y., M.M. Tabb, L. Vu, and M. Nomura. 1995. Isolation of a yeast protein kinase that is activated by the protein encoded by SRP1 (Srp1p) and phosphorylates Srp1p complexed with nuclear localization peptides. Proc. Natl. Acad. Sci. USA 92: 5159-5163.
4. Ballas, N., and V. Citovsky. 1997. Nuclear localization signal binding protein from Arabidopsis mediates nuclear import of Agrobacterium VirD2 protein. Proc. Natl. Acad. Sci., USA 94: 10723-10728.
5. Bartel, P., C. Chien, R. Sternglanz, and S. Fields. 1993. Elimination of false positives that arise in using the two-hybrid system. BioTechniques 14: 920-924.
6. Baumann, P., and S.C West. 1998. DNA end-joining catalyzed by human cell-free extracts. Proc. Natl. Acad. Sci. USA 95: 14066-14070.
7. Bravo-Angel, A.M., B. Hohn, and B. Tinland. 1997. The omega sequence of VirD2 is important but not essential for efficient transfer of T-DNA by Agrobacterium tumefaciens. Mol. Plant-Microbe Inter. 11: 57-63.
8. Broder, Y.C, A. Stanhill, N. Zakai, A. Friedler, C. Gilon, and A. Loyter. 1997. Translocation of NLS-BSA conjugates into nuclei of permeabilized mammalian cells can be supported by protoplast extract: an experimental system for studying plant cytosolic factors involved in nuclear import. FEBS Lett. 412: 535-539.
9. Burr, T.J., C. Bazzi, S. Sule, and L. Otten. 1998. Crown gall of grape: Biology of Agrobacterium vitis and the development of disease control strategies. Plant Disease 82: 1288-1297.
10. Burrell, M.M., S. Temple, and G. Ooms. 1986. Changes in translatable poly(A)RNA from differentiated potato tissues transformed with shoot-inducing Ti TL-DNA of Agrobacterium tumefaciens. Plant Mol. Biol. 6: 213-222.
11. Callis, J., M. Fromm, and V. Walbot. 1987. Introns increase gene expression in cultured maize cells. Genes. Devel. 1: 1183-1200.
12. Campos-Ortega, J.A. and V. Hartenstein. 1985. The embryonic development of Drosophila melanogaster. Berlin Heidelberg: Springer Verlag.
13. Chase, J.W. and K.R. Williams. 1986. Single-stranded DNA binding proteins required for DNA replication. Annu. Rev. Biochem. 55: 103-136.
14. Citovsky, V. 1993. Probing plasmodesmal transport with plant viruses. Plant Physiol. 102: 1071-1076.
15. Citovsky, V., G. De Vos, and P. Zambryski. 1988. Single-stranded DNA binding protein encoded by the virE locus of Agrobacterium tumefaciens. Science 240: 501-504.
16. Citovsky, V., B. Guralnick, M.N. Simon, and J.S. Wall. 1997. The molecular structure of Agrobacterium VirE2-single stranded DNA complexes involved in nuclear import. J. Mol. Biol. 271: 718-727.
17. Citovsky, V., D. Knorr, G. Schuster, and P. Zambryski. 1990. The P30 movement protein of tobacco mosaic virus is a single-strand nucleic acid binding protein. Cell 60: 637-647.
18. Citovsky, V., D. Warnick, and P. Zambryski. 1994. Nuclear import of Agrobacterium VirD2 and VirE2 proteins in maize and tobacco. Proc. Natl. Acad. Sci., USA 91: 3210-3214.
19. Citovsky, V., M.L. Wong, and P. Zambryski. 1989. Cooperative interaction of Agrobacterium VirE2 protein with single stranded DNA: Implications for the T-DNA transfer process. Proc. Natl. Acad. Sci. USA 86: 1193-1197.
20. Citovsky, V., J. Zupan, D. Warnick, and P. Zambryski. 1992. Nuclear localization of Agrobacterium VirE2 protein in plant cells. Science 256: 1802-1805.
21. Daniel, P.P. and J.A Bryant. 1988. DNA ligase in pea (Pisum sativum L.) seedlings--changes in activity during germination and effects of deoxyribonucleotides. J. Exp. Bot. 39: 481-486.
22. Davies, C., D. Howard, G. Tam, and N. Wong. 1994. Isolation of Arabidopsis thaliana mutants hypersensitive to gamma radiation. Mol. Gen. Genet. 243: 660-665.
23. de la Cruz, F. and E. Lanka. 1998. In H. Spaink, P. Hooykaas and A. Kondorosi (eds.), Function of the Ti-plasmid Vir proteins: T-complex formation and transfer to the plant cell. Kluwer Academic Press, Dordrecht, pp. 281-301.
24. Ding, L, and J.-K. Zhu. 1997. A role for arabinogalactan-proteins in root epidermal cell expansion. Planta 203: 289-294.
25. Errampalli, D., D. Patton, L. Castle, L. Mickelson, K. Hansen, J. Schnall, K. Feldmann, and D. Meinke. 1991. Embryonic lethals and T-DNA insertional mutagenesis in Arabidopsis. Plant Cell 3: 149-157.
26. Feldmann, K.A. . 1991. T-DNA insertion mutagenesis in Arabidopsis: Mutational spectrum. Plant J. 1: 71-82.
27. Feldmann, K.A. and M.D. Marks. 1987. Agrobacterium-mediated transfomation of germinating seeds of Arabidopsis thaliana: a non-tissue culture approach. Mol. Gen. Genet. 208: 1-9.
28. Fields, S. and O.-K. Song. 1989. A novel genetic system to detect protein-protein interactions. Nature 340: 245-246.
29. Filichkin, S.A. and S.B. Gelvin. 1993. Formation of a putative relaxation intermediate during T-DNA processing directed by the Agrobacterium tumefaciens VirD1, D2 endonuclease. Mol. Microbiol. 8: 915-926.
30. Fullner, K., J.C. Lara, and E.W. Nester. 1996. Pilus assembly by Agrobacterium T-DNA transfer genes. Science 273: 1107-1109.
31. Gheysen, G., R. Villarroel, and M. Van Montagu. 1991. Illegitimate recombination in plants: a model for T-DNA integration. Genes Develop. 5: 287-297.
32. Ghoshroy, S. and V Citovsky. 1998. Preservation of plant cell ultrastructure during immunolocalization of virus particles. J. Virol. Methods 74: 223-229.
33. Gietl, C., Z. Koukolikova-Nicola, and B. Hohn. 1987. Mobilization of T-DNA from Agrobacterium to plant cells involves a protein that binds single-stranded DNA. Proc. Natl. Acad. Sci. USA 84: 9006-9010.
34. Gorlich, D., and I.W. Mattaj. 1996. Nucleocytoplasmic transport. Science 271: 1513-1518.
35. Guralnick, B., G. Thomsen, and V. Citovsky. 1996. Transport of DNA into the nuclei of Xenopus oocytes by a modified VirE2 protein of Agrobacterium. Plant Cell 8: 363-373.
36. Hicks, G.R., H.M.S. Smith, S. Lobreaux, and N.V. Raikhel. 1996. Nuclear import in permeabilized protoplasts from higher plants has unique features. Plant Cell 8: 1337-1352.
37. Howard, E. and V. Citovsky. 1990. The emerging structure of the Agrobacterium T-DNA transfer complex. BioEssays 12: 103-108.
38. Howard, E.A., J.R. Zupan, V. Citovsky, and P.C. Zambryski. 1992. The VirD2 protein of A. tumefaciens contains a C-terminal bipartite nuclear localization signal: Implications for nuclear uptake of DNA in plant cells. Cell 68: 109-118.
39. Ishida, Y., H. Saito, S. Ohta, Y. Hiei, T. Komari, and T. Kumashiro. 1996. High efficiency transformation of maize (Zea mays L.) mediated by Agrobacterium tumefaciens. Nature Biotech. 14: 745-750.
40. Jasper, F., C. Koncz, J. Schell, and H.H. Steinbiss. 1994. Agrobacterium T-strand production in vitro: Sequence-specific cleavage and 5' protection of single-stranded DNA templates by purified VirD2 protein. Proc. Natl. Acad. Sci. USA 91: 694-698.
41. Kawahire, S., T. Tachibana, M. Umemoto, Y. Yoneda, N. Imai, M. Saito, T. Ichimura, S. Omata, and T. Horigome. 1996. Subcellular distribution and phosphorylation of the nuclear localization signal binding protein NBP60. Exp. Cell Res. 222: 385-394.
42. Koncz, C., N. Nemeth, G.P. Redei, and J. Schell. 1992. T-DNA insertional mutagenesis in Arabidopsis. Plant Mol. Biol. 20: 963-976.
43. Koukolikova-Nicola, Z. and B. Hohn. 1993. How does the T-DNA of Agrobacterium tumefaciens find its way into the plant cell nucleus? . Biochimie 75: 635-638.
44. Koukolikova-Nicola, Z., D. Raineri, K. Stephens, C. Ramos, B. Tinland, E.W. Nester, and B. Hohn. 1993. Genetic analysis of the virD operon of Agrobacterium tumefaciens: A search for functions involved in transport of T-DNA into the plant cell nucleus and in T-DNA integration . J. Bacteriol. 175: 723.
45. Krysan, P.J., J.C. Young, F. Tax, and M.R. Sussman. 1996. Identification of transferred DNA insertions within Arabidopsis genes involved in signal transduction and ion transport. Proc. Natl. Acad. Sci., USA 93: 8145-8150.
46. Kuldau, G.A., G. De Vos, J. Owen, G. McCaffrey, and P. Zambryski. 1990. The virB operon of Agrobacterium tumefaciens pTiC58 encodes 11 open reading frames. Mol. Gen. Genet. 221: 256-266.
47. Kussel, P and M. Frasch. 1995. Yeast Srp1, a nuclear protein related to Drosophila and mouse pendulin, is required for normal migration, division, and integrity of nuclei during mitosis. Mol. Gen. Genet. 248: 351-363.
48. Lai, E.-M. and C.I. Kado. 1998. Processed VirB2 is the major subunit of the promiscuous pilus of Agrobacterium tumefaciens. J. Bacteriol. 180: 2711-2717.
49. Laemmli, U.K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 277: 680-685.
50. Liang, P., and A.B. Pardee. 1992. Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science 257: 967-971.
51. Lister, C., and C. Dean. 1993. Recombinant inbred lines for mapping RFLP and phenotypic markers in Arabidopsis thaliana. Plant J. 4: 745-750.
52. Liu, C.-N., X.-Q. Li, and S.B. Gelvin. 1992. Multiple copies of virG enhance the transient transformation of celery, carrot, and rice tissues by Agrobacterium tumefaciens. Plant Mol. Biol. 20: 1071-1087.
53. Loeb, J.D.J., G. Schlenstedt, D. Pellman, D. Kornitzer, A. P. Silver, and G.R. Fink. 1995. The yeast nuclear import receptor is required for mitosis. Proc. Natl. Acad. Sci. USA 92: 7647-7651.
54. Marton, L., M. Hrouda, A. Pecsvaradi, and M. Czako. 1994. T-DNA-independent mutations induced in transformed plant cells during Agrobacterium co-cultivation. Transgenic Res. 3: 317-325.
55. Masson J.E., P.J. King, and J. Paszkowski. 1997. Mutants of Arabidopsis thalianahypersensitive to DNA-damaging treatments. Genetics 146: 401-407.
56. Matsumoto, S., Y. Ito, T. Hosoi, Y. Takahashi, and Y. Machida. 1990. Integration of Agrobacterium T-DNA into a tobacco chromosome: Possible involvement of DNA homology between T-DNA and plant DNA . Mol. Gen. Genet. 224: 309-316.
57. Mayerhofer, R., Z. Koncz-Kalman, C. Nawrath, G. Bakkeren, A. Cramer, K. Angelis, G.P. Redei, J. Schell, B. Hohn, and C. Koncz. 1991. T-DNA integration: A mode of illegitimate recombination in plants . EMBO J. 10: 697-704.
58. McLean, B.G., J. Zupan, and P. Zambryski. 1995. Tobacco mosaic virus movement protein associates with the cytoskeleton in tobacco cells. Plant Cell 7: 2101-2114.
59. Merkle, T., D. Lecrec, C. Marshallsay, and F. Nagy. 1996. A plant in vitro system for the nuclear import of proteins. Plant J. 10: 1177-1186.
60. Merrihew, R.V., K. Marburger, S.L Pennington, D.B. Roth, and J.H. Wilson. 1996. High-frequency illegitimate integration of transfected DNA at preintegrated target sites in a mammalian genome. Mol. Cell. Biol. 16: 10-18.
61. Mindrinos, M., F. Katagiri, G.-L. Yu, and F.M. Ausubel. 1994. The A. thaliana disease resistance gene RPS2 encodes a protein containing a nucleotide-binding site and leucine-rich repeats. Cell 78: 1089-1099.
62. Minet, M., M.E. Dufour, and F. Lacroute. 1992. Complementation of Saccharomyces cervisiae auxotrophic mutants by Arabidopsis thaliana cDNAs. Plant J. 2: 417-422.
63. Mysore, K.S., B. Bassuner, X.-b. Deng, N.S. Darbinian, A. Motchoulski, W. Ream, and S.B. Gelvin. 1998. Role of the Agrobacterium tumefaciens VirD2 protein in T-DNA transfer and integration. Mol. Plant-Microbe Inter. 11: 668-683.
64. Nam, J., A.G. Matthysse, and S.B. Gelvin. 1997. Differences in susceptibility of Arabidopsis ecotypes to crown gall disease may result from a deficiency in T-DNA integration. Plant Cell 9: 317-333.
65. Nam, J., K.S. Mysore, C. Zheng, M. Knue, A.G. Matthysse, and S.B Gelvin. 1999. Identification of T-DNA tagged Arabidopsis mutants that are resistant to Agrobacterium transformation. Mol. Gen. Genet. In press.
66. Napoli, C., C. Lemieux, and R. Jorgensen. 1990. Introduction of a chimeric chalcone synthase gene into Petunia results in reversible co-suppression of homologous genes in trans. Plant Cell 2: 279-289.
67. Narasimhulu, S.B., X.-B. Deng, R. Sarria, and S.B. Gelvin. 1996. Early transcription of Agrobacterium T-DNA genes in tobacco and maize. Plant Cell 8: 873-886.
68. Ni, M., D. Cui, J. Einstein, S. Narasimhulu, C.E. Vergara, and S.B. Gelvin. 1995. Strength and tissue specificity of chimeric promoters derived from the octopine and mannopine synthase genes. Plant J. 7: 661-676.
69. Offringa, R., M.J. de Groot, H.J. Haagsman, M.P. Does, P.J. van den Elzen, and P.J. Hooykaas. 1990. Extrachromosomal homologous recombination and gene targeting in plant cells after Agrobacterium mediated transformation. EMBO J. 9: 3077-3084.
70. Ohba, T., Y. Yoshioka, C. Machida, and Y. Machida. 1995. DNA rearrangement associated with the integration of T-DNA in tobacco: An example for multiple duplications of DNA around the integration target. Plant J. 7: 157-164.
71. Otten, L., H. DeGreve, J. Leemans, R. Hain, P. Hooykaas, and J. Schell. 1984. Restoration of virulence of vir region mutants of Agrobacterium tumefaciens strain B6S3 by coinfection with normal and mutant Agrobacterium strains. Mol. Gen. Genet. 195: 159-163.
72. Pansegrau, W., F. Schoumacher, B. Hohn, and E. Lanka. 1993. Site-specific cleavage and joining of single-stranded DNA by VirD2 protein of Agrobacterium tumefaciens Ti plasmids: Analogy to bacterial conjugation. Proc. Natl. Acad. Sci. USA 90: 11538-11542.
73. Paszkowski, J., M. Baur, A. Bogucki, and I. Potrykus. 1988. Gene targeting in plants. EMBO J. 7: 4021-4026.
74. Pear, J.R., Y. Kawagoe, W.E. Schreckengost, D.P. Delmer, and D.M. Stalker. 1996. Higher plants contain homologs of the bacterial celA genes encoding the catalytic subunit of cellulose synthase. Proc. Nat. Acad. Sci. USA 93: 12637-12642.
75. Regensburg-Tuink, A.J.G. and P.J.J. Hooykaas. 1993. Transgenic N. glauca plants expressing bacterial virulence gene virF are converted into hosts for nopaline strains of A. tumefaciens. Nature 363: 69-71.
76. Reiter, R.S., J.G.K. Williams, K.A. Feldmann, J.A. Rafalski, S.V. Tingey, and P.A.Scolnik. 1992. Global and local genome mapping in Arabidopsis thaliana by using recombinant inbred lines and random amplified polymorphic DNAs. Proc. Natl. Acad. Sci. USA 89: 1477-1481.
77. Relic, B., M. Andjelkovic, L. Rossi, Y. Nagamine, and B. Hohn. 1998. Interaction of the DNA modifying proteins VirD1 and VirD2 of Agrobacterium tumefaciens: Analysis by subcellular localization in mammalian cells. Proc. Natl. Acad. Sci., USA 95: 9105-9110.
78. Ritchie, S.W., C.-N. Liu, J.C. Sellmer, H. Kononowicz, T.K. Hodges, and S.B. Gelvin. 1993. Agrobacterium tumefaciens-mediated expression of gusA in maize tissues. Transgenic Res. 2: 252-265.
79. Roe, J.L, C.J. Rivin, R.A. Sessions, K.A. Feldmann, and P. Zambryski. 1993. The tousled gene in A. thaliana encodes a protein kinase homolog that is required for leaf and flower development. Cell 75: 939-950.
80. Rong, L., S.J. Karcher, and S.B. Gelvin. 1991. Genetic and molecular analyses of picA, a plant-inducible locus on the Agrobacterium tumefaciens chromosome. J. Bacteriol. 173: 5110-5120.
81. Rong, L., S.J. Karcher, K. O'Neal, M.C. Hawes, C.D. Yerkes, R.K. Jayaswal, C.A. Hallberg, and S.B. Gelvin. 1990. picA, a novel plant-inducible locus on the Agrobacterium tumefaciens chromosome. J. Bacteriol. 172: 5828-5836.
82. Rossi, L., B. Hohn, and B. Tinland. 1993. The VirD2 protein of Agrobacterium tumefaciens carries nuclear localization signals important for transfer of T-DNA to plants . Mol. Gen. Genet. 239: 345.
83. Rossi, L., B. Hohn, and B. Tinland. 1996. Integration of complete transferred DNA units is dependent on the activity of virulence E2 protein of Agrobacterium tumefaciens. Proc. Natl. Acad. Sci., USA 93: 126-130.
84. Ruan, Y., J. Gilmore, and T. Conner. 1998. Towards Arabidopsis genome analysis: Monitoring expression profiles of 1400 genes using cDNA microarrays. Plant J. 15: 821-833.
85. Salomon, S. and H. Puchta. 1998. Capture of genomic and T-DNA sequences during double-strand break repair in somatic cells. EMBO J. 17: 6086-6095.
86. Schlenstedt, G., E. Hurt, V. Doye, and P. Silver. 1993. Reconstitutuion of nuclear protein transport with semi-intact yeast cells. J. Cell Biol. 123: 785-798.
87. Schrammeijer, B., J. Hemelaar, and P.J.J. Hooykaas. 1998. The presence and characterization of a virF gene on Agrobacterium vitis Ti plasmids. Mol. Plant-Microbe Inter. 11: 429-433.
88. Sen, P., G. J. Pazour, D. Anderson, and A. Das. 1989. Cooperative binding of Agrobacterium tumefaciens VirE2 protein to single-stranded DNA. J. Bacteriol. 171: 2573-80.
89. Shen, W.-H., J. Escudero, M Schlappi, C. Ramos, B. Hohn, and Z. Koukolikova-Nicola. 1993. T-DNA transfer to maize cells: Histochemical investigation of b-glucuronidase activity in maize tissues. Proc. Natl. Acad. Sci. USA 90: 1488-1492.
90. Shen, W.H. and C. Gigot. 1997. Protein complexes binding to cis elements of the plant histone gene promoters: multiplicity, phosphorylation and cell cycle alteration. Plant Mol. Biol. 33: 367-379.
91. Sheng, J. and V. Citovsky. 1996. Agrobacterium-plant cell DNA transport: Have virulence proteins, will travel. Plant Cell 8: 1699-1710.
92. Shirasu, K. and C.I. Kado. 1993. Membrane location of the Ti plasmid VirB proteins involved in the biosynthesis of a pilin-like conjugative structure on Agrobacterium tumefaciens. FEMS Microbiol. Lett. 111: 287-294.
93. Shurvinton, C.E., L. Hodges, and W. Ream. 1992. A nuclear localization signal and the C-terminal omega sequence in the Agrobacterium tuemfaciens VirD2 endonuclease are important for tumor formation. Proc. Natl. Acad. Sci., USA 89: 11837-11841.
94. Stachel, S.E., B. Timmerman, and P. Zambryski. 1986. Generation of single-stranded T-DNA molecules during the initial stages of T-DNA transfer from Agrobacterium tumefaciens to plant cells. Nature (London) 322: 706-712.
95. Thompson, D.V., L.S. Melchers, K.B. Idler, R.A. Schilperoort, and P.J.J. Hooykaas. 1988. Analysis of the complete nucleotide sequence of the Agrobacterium tumefaciens virB operon. Nuc. Acids Res. 16: 4621-4636.
96. Tinland, B., Z. Koukolikova-Nicola, M.N. Hall, and B. Hohn. 1992. The T-DNA-linked VirD2 protein contains two distinct functional nuclear localization signals . Proc. Natl. Acad. Sci., USA 89: 7442-7446.
97. Tinland, B., F. Schoumacher, V. Gloeckler, A.M. Bravo-Angel, and B. Hohn. 1995. The Agrobacterium tumefaciens virulence D2 protein is responsible for precise integration of T-DNA into the plant genome. EMBO J. 14: 3585-3595.
98. Tomkinson, A.E. and D.S. Levin. 1997. Mammalian DNA ligases. BioEssays 19: 893-901.
99. Van Lijsebettens, M., R. Vanderhaeghen, and M. Van Montagu. 1991. Insertional mutagenesis in Arabidopsis thaliana: Isolation ofa T-DNA linked mutation that alters leaf morphology. Theor. Appl. Genet. 2: 277-284.
100. Wada, T., T. Tachibana, Y. Shimura, and K. Okada. 1997. Epidermal cell differentiation in Arabidopsis determined by a Myb homolog, CPC. Science 277: 1113-1116.
101. Ward, J.E., D.E. Akiyoshi, D. Regier, A. Datta, M.P. Gordon, and E.W. Nester. 1988. Characterization of the virB operon from an Agrobacterium tumefaciens Ti plasmid . J. Biol. Chem. 263: 5804-5814.
102. Weis, K., I. W. Mattaj, and A.I. Lamond. 1995. Identification of hSRP1a as a functional receptor for nuclear localization sequences. Science 268: 1049-1053.
103. Winans, S.C. . 1992. Two-way chemical signaling in Agrobacterium-plant interactions. Microbiol. Rev. 56: 12-31.
104. Yusibov, V.M., T.R. Steck, V. Gupta, and S.B. Gelvin. 1994. Association of single-stranded transferred DNA from Agrobacterium tumefaciens with tobacco cells. Proc. Natl. Acad. Sci., USA 91: 2994-2998.
105. Zambryski, P.C. 1992. Chronicles from the Agrobacterium-plant cell DNA transfer story. Annu. Rev. Plant Physiol. Plant Mol. Biol. 43: 465-490.
106. Ziemienowicz A., B. Tinland, J. Bryant, V. Gloeckler, and B. Hohn. Plant enzymes but not Agrobacterium VirD2 mediate T-DNA ligation in vitro. In preparation.
107. Zupan, J.R., V. Citovsky, and P. Zambryski. 1996. Agrobacterium VirE2 protein mediates nuclear uptake of single-stranded DNA in plant cells. Proc. Natl. Acad. Sci., USA 93: 2392-2397.
108. Zupan, J.R., and Zambryski, P. 1995. Transfer of T-DNA from Agrobacterium to the plant cell. Plant Physiol. 107: 1041-1047.
109. Zupan, J., and P. Zambryski. 1997. The Agrobacterium DNA transfer complex. Crit. Rev. Plant Science16: 279-295.
(Return to top of document)
(A-1) THE PROPOSED PROJECT AND CURRENT ACTIVITIES
Vitaly Citovsky
A. Current Activities
My laboratory uses plant systems to study two basic biological processes: (i) nuclear import and (ii) virus-host interactions.
I. Transport of Agrobacterium T-DNA into the host cell nucleus as an experimental model for plant nuclear import. Nuclear traffic of macromolecules is a basic cellular process in all eukaryotic organisms. We study nuclear uptake in plants using Agrobacterium-plant cell interaction during which bacterial T-DNA-protein complexes (T-complexes) are imported into the host cell nucleus. Since pathogenic microorganisms often adapt existing cellular machinery for their own needs, Agrobacterium likely employs an endogenous cellular pathway for nuclear import and, thus, represents a convenient experimental system for nuclear transport of both nucleic acid and protein molecules. Our studies have identified two Agrobacterium proteins, VirD2 and VirE2, which form a complex with the transported T-DNA and target it to and through the nuclear pore. We have also shown that while VirD2 localizes to the cell nucleus both in plant and animal systems, the nuclear targeting activity of VirE2 is plant-specific. Rearrangement of a single amino acid residue in this plant-specific nuclear localization signal (NLS) of VirE2 enables it to function in animal cells, efficiently delivering DNA molecules into their nuclei. Currently, we are identifying and characterizing host plant proteins that interact with Agrobacterium VirD2 and VirE2. For example, we have cloned an Arabidopsis gene coding for an NLS-binding protein, AtKAPa, which is directly involved in VirD2 nuclear import.
In addition to the Agrobacterium T-complex nuclear import, we are investigating a specific case of nuclear transport of the tomato yellow leaf curl geminivirus (TYLCV) which is a major pathogen of tomato plants. In international collaboration with Dr. Gafni (Volcani Center, Israel), we have shown that TYLCV coat protein carries a functional NLS and is likely involved in nuclear targeting of the viral genomic DNA.
II. Plant-virus interactions: Viral cell-to-cell and systemic movement and development of viral disease. Plant intercellular communication largely occurs via cell-to-cell connections, the plasmodesmata (PD). As an experimental system to study PD transport and regulation, we are using cell-to-cell movement proteins of plant viruses as which interact with PD to increase their size exclusion limit. Thus, these proteins are utilized as tools to identify their cellular receptors involved in PD transport. In addition, we have isolated a mutant of Arabidopsis thaliana, a genetic model plant, which is defective in viral systemic movement and, by implication, in PD transport. This mutant, designated vsm1, is used to elucidate the molecular pathways for viral systemic spread. Finally, we are isolating Arabidopsis mutants with alterations in viral disease symptoms. One such mutant, vid1, is indistinguishable from the wild-type plants when healthy but develops a severely dwarfed phenotype with loss of apical dominance when infected. The study of vid1 plants may help to better understand the mechanisms of formation of plant viral disease symptoms.
B. Relationship to the Proposed Project
There is a "feedback" relationship between the proposed project and my current research. First, the proposal is directly related to my ongoing T-DNA nuclear import project, benefiting from our experience in this field. The proposed research builds on my long-term experience in the biochemical, molecular, and cellular aspects of DNA transfer from Agrobacterium to plant cells. In addition, the proposed project takes advantage of my general expertise in plant-pathogen interactions, such as identification of Arabidopsis mutants with altered symptoms of viral disease and/or resistance to viral systemic movement. Our work on these projects has provided my research group with experience in confocal and electron microscopy, microinjection, yeast two-hybrid system, in vitro nuclear import, protein-DNA interaction, and genetic screening for Arabidopsis mutants and mapping of the identified mutations. These approaches also represent the major technical tools for the proposed research.
Second, the proposed collaborative project will in turn enhance my entire research program. It will provide new insights into the molecular mechanisms of Agrobacterium infection which most likely may be applied to other cases of plant-pathogen interactions, such as viral infection, another major focus of my current research.
Stanton B. Gelvin
A. Current Activities
My laboratory studies the initial events of Agrobacterium-plant interactions, including T-DNA and VirE2 transport from the bacterium, bacterial attachment to the plant cell, nuclear targeting of the T-complex, and T-DNA integration into the plant genome. Also, we are examining the use of Agrobacterium-mediated transformation for homologous and site-directed insertion of T-DNA into the plant genome, and the activity of novel "artificial" promoters constructed using T-DNA promoters and transcriptional activating elements.
I. Mechanism of oncogenic suppression of Agrobacterium by the plasmid pSa. When coresidient with the Ti-plasmid in Agrobacterium, the IncW plasmid pSa inhibits tumorigenesis (oncogenic suppression, OS). A single gene of pSa, osa (oncogenic suppressive activity) is sufficient to effect oncogenic suppression. We have recently determined that the molecular mechanism of OS involves inhibition by osa of VirE2 export from Agrobacterium, but not T-DNA export. In addition, pSa can inhibit the conjugal transfer of the IncQ plasmid RSF1010 between Agrobacterium cells. Our data indicate that osa interacts with the VirB/VirD4 transport apparatus to inhibit conjugal transfer (fertility inhibition) and to cause OS. We are determining the component of the this transport apparatus with which Osa protein interacts.
II. Identification of plant proteins that interact with Vir protein nuclear transport signals, and the regulation of T-complex nuclear transport. We have used a yeast two-hybrid system to identify a plant type 2C protein phosphatase (PP2C) that interacts specifically with the nuclear localization signal (NLS) of VirD2 protein. An Arabidopsis PP2C mutant (abi1) shows increased levels of Agrobacterium transformation. This PP2C negatively regulates nuclear import of VirD2 protein, and hence the T-complex, by dephosphorylating a serine residue adjacent to the VirD2 NLS. Alteration of this serine residue to an alanine decreases VirD2 nuclear import and transformation of plant cells by Agrobacterium. We are currently investigating the kinase that is responsible for phosphorylation of this serine, and thus potentiating T-complex nuclear transport.
III. Determination of the relative contributions of VirD2 and VirE2 NLSs to T-complex nuclear transport. We can generate "artificial T-complexes" by cleaving fluorescently-labeled single-stranded DNA molecules containing a T-DNA border with VirD2 protein in vitro. These DNA/VirD2 complexes can be coated with VirE2 protein in vitro. By microinjecting fluorescently-labeled T-complexes containing wild-type or mutant VirD2 NLSs into plant cells, we are currently measuring the relative contributions of the VirD2 and the VirE2 NLSs to the rate and extent of T-complex nuclear import.
IV. Identification of Arabidopsis mutants and genes involved in Agrobacterium transformation. We are currently screening various T-DNA tagged Arabidopsis collections for rat (resistant to Agrobacterium transformation) mutants. Most of the currently-identified mutants are blocked at an early stage of the transformation process (i.e., bacterial attachment, T-complex transport to the plant, or T-complex nuclear transport). Some mutants are blocked in T-DNA integration into the plant genome. Isolation of T-DNA/plant DNA junction fragments and the corresponding Arabidopsis cDNA and genomic clones has resulted in the identification of several plant genes involved in Agrobacterium attachment (an arabinogalactan protein, a cellulose synthase-like protein, and a wall protein) and in T-DNA integration (a histone H2A protein and a myb-like transcription factor).
V. Generation of novel promoters useful for plant genetic engineering. We have constructed a strong promoter (the "super-promoter") from transcriptional regulatory elements of the octopine synthase and mannopine synthase 2' genes. We are currently concluding an analyis of the strengths of this promoter in monocots and dicot plant species.
VI. Use of Agrobacterium to effect homologous and site-directed recombination in plants. We are using Agrobacterium vir mutants that decrease illegitimate recombination of the T-DNA with the plant genome to effect more efficient homologous and site-directed recombination in plants.
B. Relationship to the Proposed Project
The major current goal of research in my laboratory is to identify plant components necessary for Agrobacterium-mediated transformation, and to utilize this information to genetically manipulate currently recalcitrant plant species so that they can more readily be transformed. The proposed project greatly enhances my capabilities to achieve these goals by providing the resources (both financial and the benefits of a collaboration with experts in the field) to extend these investigations. Studies of plant genes regulated during the early stages of transformation and plant proteins that interact with Vir proteins will provide information to perform reverse genetic analysis of mutant plants to determine the roles of these genes and proteins in the transformation process. The identification of rat mutants will complement these efforts by directly providing information about specific genes and their functions in transformation.
Barbara Hohn
A. Current Activities
I. Transfer and Integration of T-DNA. Interaction of Agrobacterium with plants is a fascinating research topic as a whole, yet we now concentrate on work analyzing the fate of T-DNA in the plant. The specific steps currently worked on are import of T-DNA into plant nuclei and integration of T-DNA. Precise integration of T-DNA is dependent on both virulence proteins VirD2 and VirE2. In the absence of functional versions of these proteins transfer is inefficient, nuclear import is severely reduced and integration of a low level of truncated versions of T-DNA is detected only. Current activities focus on in vitro analysis of T-DNA integration, using model targets and model T-DNAs.
II. Homologous recombination in plants. Using transgenic Arabidopsis thaliana and tobacco plants which allow quantitative determination of intrachromosomal homologous recombination we analyze influences of known mutations in various pathways on recombination. Also environmental effects, biotic and abiotic in nature, on homologous recombination are being studied.
B. Relationship to the Proposed Project
The proposed work will be of mutual benefit. The planned collaborative research will profit from my long-term experience in Agrobacterium-plant interaction on a mostly molecular-biological level. Likewise, homologous recombination is an area new for my prospective collaborators and the project integrates this work. Constructs, assays, plants and an enthusiastic personal environment (over the distance or in our institute) will be available to anybody in the project.
Conversely, the projected work, being to a good proportion a direct extension of ongoing work at our laboratory in Basel, will provide new dimensions in our thinking on Agrobacterium and plants.
(Return to top of document)
(A-2) INTELLECTUAL PROPERTY
None of the data, bacterial strains, plasmids, genes, or mutants that we generate during the course of this project will be considered proprietary. We shall immediately deposit all information obtained during the course of this work in the appropriate data bases (e.g., GeneBank, Arabidopsis mutant data base, or the web site that we shall establish). We shall freely distribute all data, bacterial strains, plasmids, genes, mutants, etc. generated during the course of this work.
(A-3) MANAGEMENT PLAN
A. Management and Collaboration
The logistics of the proposed collaboration is clear. It will be performed on three levels. First, all three labs will be in constant electronic communication to exchange up-to-date information on the ongoing experiments. Each time a significant discovery is made (e.g. a new host factor or biological activity is identified), we will arrange for a three-way telephone conference calls to interpret the experimental results, plan our subsequent experiments, and prepare publications and presentations of the obtained data. Once a year, the PI and two co-PIs will personally meet in one of the labs for extensive and comprehensive discussions and planning.
Second, each laboratory will be assigned a specific task of conducting experimentation in its area of expertise. Arabidopsis genes, proteins, and mutant plants will be shipped from the lab in which they had been identified to the designated lab best suited to assay their biological function. For example, VirD2- and VirE2-interacting proteins isolated in the Citovsky lab will be shipped to the Gelvin lab to be tested for their role in nuclear import in plant protoplasts and to the Hohn lab to be tested for their involvement in T-DNA integration. Or, rat mutants deficient for transient T-DNA expression identified in the Gelvin lab will be sent to the Citovsky lab where they will be examined for the presence of AtKAPalpha and other factors required for VirD2 and VirE2 nuclear import.
Third, in case of especially laborious experiments, several laboratories will share the same experimental load. For example, both Gelvin and Citovsky labs are experienced in genetic approaches to Arabidopsis-pathogen interactions; thus, the genetic screening rat mutants and subsequent cloning of the T-DNA tagged RAT genes will be performed in both laboratories simultaneously. To further facilitate the exchange of laboratory expertise, postdoctoral associates from each group will travel to other sister-labs to perform collaborative experiments and learn new experimental approaches.
B. Training And Diversity
State University of New York at Stony Brook
The Molecular and Cellular Biology (MCB) Training Program run by the Department of Biochemistry and Cell Biology is the major multidisciplinary, inter-departmental graduate program in the biological sciences at SUNY, Stony Brook. The Program is supported by the NIH Training Grant #GM08468 and includes faculty members from Stony Brook, Cold Spring Harbor, and Brookhaven National Laboratory. Thus, the MCB graduate students have access to the intellectual resources and infrastructural facilities of these three leading research institutions. On average, the MCB program receives between 250-300 applications a year which result in an entering class of 20 students. A notable aspect of this recruitment is the increased success of minority recruiting. Last year, for example, 8 minority students were interviewed, resulting in extending offers to 4 of these students.
To enhance further minority recruitment, the MCB program wrote a proposal for federal Fellowships (Title IX-B) for underrepresented minority students and was one of only three programs on campus to receive a Patricia Roberts Harris Fellowship for an incoming student in The Program. The MCB program also is represented at the NIGMS Minority Programs Symposium in Washington, D.C. The MCB program has established a relationship with the Minority Apprenticeship Program (M-RAP). During summers, several M-RAP students, who are biology/biochemistry undergraduates in other institutions, work in MCB members' laboratories. Also, the MCB program has close ties to the Biology department of Hunter College of the City University of New York which is the focus of the minority students of the CUNY system. These relationships with M-RAP and CUNY students allow the MCB program to attract and recruit the minority students from nationwide as well as from the New York Metropolitan area. Finally, our we routinely solicit applications from the students participating in the MBRS and MARC programs and students listed on the Minority Locator Service.
All of the MCB minority students are recipients of W. Burghart Turner Graduate Fellowships awarded competitively by the State of New York. In addition, one of the MCB minority students was recently awarded a Howard Hughes Predoctoral Fellowship, further attesting to the high quality of minority students in the Program. Besides W. Burghart Turner and Howard Hughes Predoctoral Fellowships, the MCB graduate students can apply for an Institute of Cell and Developmental Biology (ICDB) Predoctoral Fellowship which is funded by pharmaceutical and biotech firms.
In addition to this major effort by the MCB program to recruit minority graduate students, the co-PI (VC) himself is actively seeking postdoctoral trainees from the underrepresented ethnic/racial groups. For example, Dr. Robert Lartey, an African-American, was successfully recruited to join this co-PI's lab. During his three-year postdoctoral training, Dr. Lartey published in leading plant biology journals and was supported by a minority postdoctoral grant from the NIH.
Once the minority graduate students and postdocs are recruited to the co-PI's lab at Stony Brook, they take advantage of a very stimulating environment with faculty interests in such different fields as plant biology, neurobiology, Drosophila, Xenopus and mouse development, and yeast biology. In addition to the departmental structure, the co-PI's laboratory is a member of ICDB which includes a recently-awarded Howard Hughes funding. Both the Department and ICDB run weekly series of seminars that host prominent scientists from the US and abroad, including Nobel laureates and National Academy members. Importantly, plant biology represents a frequent topic of these lectures. In addition to the formal seminars and numerous journal clubs, ICDB supports an annual three day-long retreat in Shelter Island, NY, where faculty, postdocs and graduate students give informal talks and poster presentations, further fostering interaction and collaboration between labs in different fields of biology. Lastly, the proximity of Cold Spring Harbor Laboratory, which is a member of our graduate program, allows attendance of various courses and workshops as well as easy interaction with the CSH staff.
Purdue University
Dr. Gelvin is a member of Department of Biological Sciences and the Interdisciplinary programs Biochemistry and Molecular Biology (BMB), Purdue Genetics Program (PGP), and Plant Biology Program (PBP). Each of these departments/programs has a strong plant biology component, including a large number of faculty who specialize in the area of plant genomics, and have consistently recruited excellent graduate students and postdoctoral research fellows in the area of plant biology. Students (and postdoctoral fellows) in these programs have the opportunity of enrolling in numerous courses in basic plant sciences, including plant molecular biology (taught by SBG), plant cell biology, plant growth and development, plant genetics and genomics, plant biochemistry, and numerous specialty courses in the applied and basic plant sciences. In addition to numerous journal clubs in various life science disciplines, including plant sciences, a group of approximately 10 plant cell and molecular biology laboratories (including SBG) holds weekly research conferences to acquaint members with ongoing research in each of the participating laboratories. These meetings are attended by 20-40 graduate students, postdoctoral fellows, and research advisors. Many of these students are supported by a NSF Plant Genetics Training Grant or a USDA National Needs Training Grant. In addition to the numerous weekly seminar series in the life sciences, students and postdoctoral fellows are encouraged to attend the weekly Plant Cell and Molecular Biology Seminar series (established approximately 15 years ago). This seminar series brings national and international plant scientists to the campus, and first year students in the PBP are required to participate in a "student seminar course" revolving around this weekly seminar series.
Special efforts have been and continue to be made to recruit women and minority students and postdoctoral research fellows. Of the 436 applicants to the Department of Biological Sciences in 1997-1998, 37 (8.5%) were black, hispanic, or Asian/Pacific Island students. Of the currently enrolled 122 students in the department, 16 (13%) represent minority groups. These students are supported by NIH fellowships, a Biophysics Training Grant, Purdue University minority fellowships, and special Abbott, Genox, Indiana Elks Club, and Sloan Foundation Fellowships. 42% of the current Biology Ph.D. students are female. Over the past 5 years, the BMB program has enrolled 8 (of 74) minority students, and the smaller PGP and PBP programs enrolled 2 minority students (of 11 total students) in 1998. Purdue University maintains an active minority recruiting effort, including participation in summer MARC and AIM undergraduate research programs (SBG has participated in the MARC program, and successfully recruited a MARC student to his laboratory), and sending faculty to Traditional Minority Colleges to recruit graduate students. SBG has recently participated in this program and has recruited students from Tuskegee University.
(A-4) COORDINATION WITH OUTSIDE GROUPS
This project is not part of another national or international collaborative project.
(Return to top of document)
This work is made possible by a grant from the National
Science Foundation's Plant Genome Research Program.
For additional information about this research, contact:
Dr. Stanton Gelvin (gelvin@bilbo.bio.purdue.edu)
765/494-4939
Department of Biological Sciences
Purdue University
1392 Lilly Hall
West Lafayette, IN 47907-1392
Questions or Comments regarding this web site should be addressed to Dr. Stanton Gelvin
![]()



